June 10, 2014
Filtering EST and mRNA BLAST results to get final figures-- breaking up each table into 3 aspects (cellular components, biological processes, and molecular function)EST taxID6580 all results
link to BLAST RESULTS
mRNA-- http://goo.gl/JrJDzlESTs-- http://goo.gl/WWQXmA
pie GoSlim terms-- biological process
should be about 13
May 30, 2014
qPCR of geoduck larvae| Well/rep |
Sample |
Stage |
Primer Set |
Developmental Process |
|
| A1/B1 |
T1 |
Trocophore |
Rp_RNF66_F |
Rp_RNF67_R |
multicellular organismal development |
| A2/B2 |
T2 |
Trocophore |
|||
| A3/B3 |
V1 |
Veliger |
|||
| A4/B4 |
V2 |
Veliger |
|||
| A5/B5 |
S1 |
Settler |
|||
| A6/B6 |
S2 |
Settler |
|||
| A7/B7 |
NTC |
-- |
|||
| C1/D1 |
T1 |
Trocophore |
Mm_flilCG1484_F |
Mm_flilCG1485_R |
nematode larval development |
| C2/D2 |
T2 |
Trocophore |
|||
| C3/D3 |
V1 |
Veliger |
|||
| C4/D4 |
V2 |
Veliger |
|||
| C5/D5 |
S1 |
Settler |
|||
| C6/D6 |
S2 |
Settler |
|||
| C7/D7 |
NTC |
-- |
|||
| Rp_RNF66/67 |
Mm_flilCG1484/5 |
||||||
| [Stock] |
ul/rxn |
Cocktail |
'# of NTC |
ul/rxn |
Cocktail |
'# of NTC |
|
| Water |
7 |
98 |
2 |
7 |
98 |
2 |
|
| Primer - F (uM) |
10 |
0.5 |
7 |
'# of Unknowns |
0.5 |
7 |
'#of Unknowns |
| Primer - R (uM) |
10 |
0.5 |
7 |
6 |
0.5 |
7 |
6 |
| Sso Fast EvaGreen Supermix (2X) |
1X |
10 |
140 |
Replicate |
10 |
140 |
Replicate |
| 2 |
2 |
||||||
| cDNA |
2 |
Total # Samples |
2 |
Total # Samples |
|||
| Total volume (ul) |
20 |
20 |
280 |
14 |
20 |
280 |
14 |
Results:
Larvae qPCR output
| PLATE SET UP |
|||||||
| 1 |
2 |
3 |
4 |
5 |
6 |
7 |
|
| A |
T1 Rp |
T2 Rp |
V1 Rp |
V2 Rp |
S1 Rp |
S2 Rp |
NTC |
| B |
T1 Rp |
T2 Rp |
V1 Rp |
V2 Rp |
S1 Rp |
S2 Rp |
NTC |
| C |
T1 Mm |
T2 Mm |
V1 Mm |
V2 Mm |
S1 Mm |
S2 Mm |
NTC |
| D |
T1 Mm |
T2 Mm |
V1 Mm |
V2 Mm |
S1 Mm |
S2 Mm |
NTC |
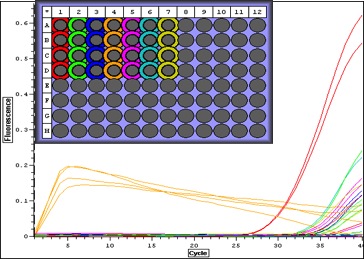 |
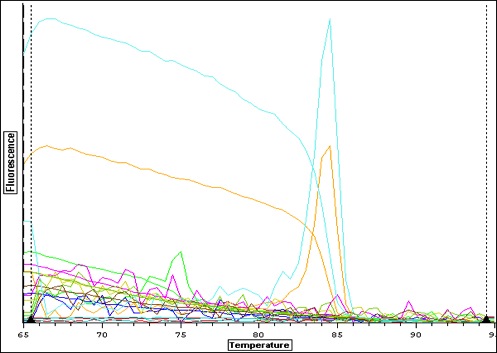
| Quant. plot for all samples. Peak for V2 (column 4) may have something to do with amount of RNA-- notes in Sam's notebook that during RNA extraction there was a "goo" and the yield was very high. |
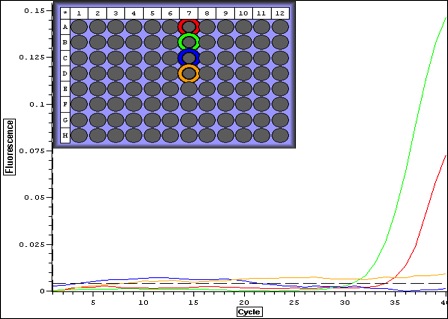 |
| Quant plot for Negative controls only. May be contamination.. which means that maybe false positives for larvae. |
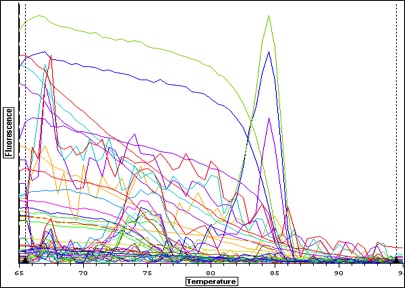 |
| Melt curve for all samples-- plate colors need to be re-uploaded from original file-- I think I copied over the wrong plate layout-- colors don't match above. |
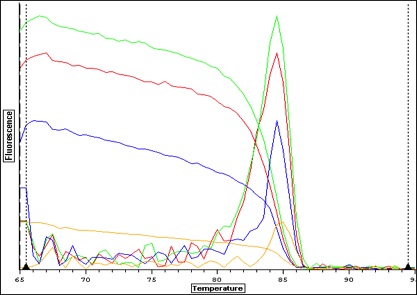 |
| Melt curve for positive signals with one solid peak-- corresponding wells are depicted in plate picture below |
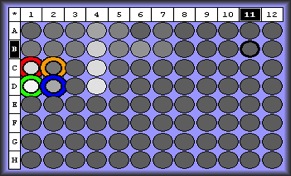
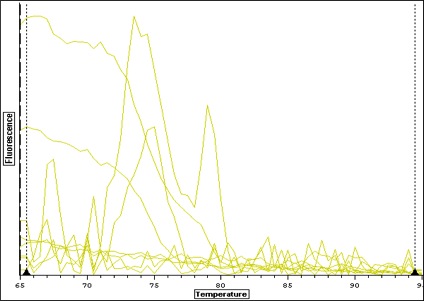 |
| Melt curve for negative controls--- need to ask Steven how to interpret this.. looks like not one product so maybe not contamination that would make a false positive? |
May 29, 2014
qPCR Results: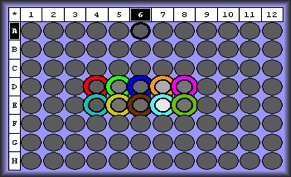
| Well |
Working label |
Primer Set |
Developmental Process |
|
| D4/E4 |
NTC |
N/A |
N/A |
N/A |
| D5/E5 |
3 |
Rp_RNF66_F |
Rp_RNF67_R |
multicellular organismal development |
| D6/E6 |
4 |
Rp_flilCG1484_F |
Rp_flilCG1485_R |
muscle fiber development |
| D7/E7 |
5 |
Mm_flilCG1484_F |
Mm_flilCG1485_R |
nematode larval development |
| D8/E8 |
9 |
Mm_HCA57_F |
Mm_HCA57_R |
embryonic development ending in birth or egg hatching |
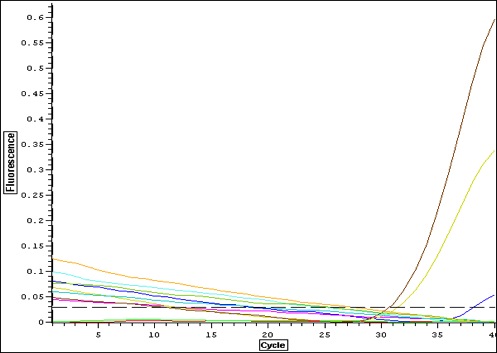
Quantification Output
| Well / Set |
Dye |
Content |
Description |
Efficiency |
C(t) |
Quantity |
Avg C(t) |
| D4 |
SBGnew |
Sample |
NTC |
N/A |
N/A |
||
| D5 |
SBGnew |
Sample |
Rp_RNF66/7 |
1.48 |
38.13 |
||
| D6 |
SBGnew |
Sample |
Rp_flilCG1484/5 |
N/A |
N/A |
||
| D7 |
SBGnew |
Sample |
Mm_flilCG1484/5 |
1.77 |
31.44 |
||
| D8 |
SBGnew |
Sample |
Mm_HCA57 |
N/A |
N/A |
||
| E4 |
SBGnew |
Sample |
NTC |
N/A |
N/A |
||
| E5 |
SBGnew |
Sample |
Rp_RNF66/7 |
N/A |
N/A |
||
| E6 |
SBGnew |
Sample |
Rp_flilCG1484/5 |
N/A |
N/A |
||
| E7 |
SBGnew |
Sample |
Mm_flilCG1484/5 |
1.97 |
30.68 |
||
| E8 |
SBGnew |
Sample |
Mm_HCA57 |
N/A |
N/A |
||
selecting primers 3 and 5 to apply to larval stages. Mm_flilCG1484/5 appears to give a strong signal with one solid product. Primer 3 only had one replicate amplify-- will apply to larvae anyway.
Made cDNA out of RNA extracts Sam White isolated 5/27-5/28
6 samples total:
T1, T2 (trocophores)
V1, V2 (veligers)
S1, S2 (setters interphase)
| Min. Per Rxn (ul) |
Total / Sample (ul) |
|
| RNA |
17.75 |
35.5 |
| oligo dT |
0.5 |
1 |
| 70C for 5 min |
||
| Ice |
||
| Master Mix |
||
| Per Rxn (ul) |
Total (ul) |
|
| M-MLV RT buffer (5X) |
5 |
60 |
| dNTPs (10mM) |
1.25 |
15 |
| M-MLV RT |
0.5 |
6 |
May 27, 2014
| [Stock] |
ul/rxn |
Cocktail (per primer) |
Total Volume |
-# of NTC |
|
| Water |
10 |
20 |
100 |
2 |
|
| Primer - F (uM) |
10 |
0.5 |
1 |
5 |
-#of Unknowns |
| Primer - R (uM) |
10 |
0.5 |
1 |
5 |
4 |
| Sso Fast EvaGreen Supermix (2X) |
1X |
10 |
20 |
100 |
Replicate |
| 2 |
|||||
| cDNA |
2 |
4 |
20 |
Total # Samples |
|
| Total volume (ul) |
20 |
23 |
46 |
230 |
10 |
| Thermalcycler conditions |
Cycles |
||||
| 95 C |
30 sec |
1 |
|||
| 95 C |
5 sec |
30-40 |
|||
| 55-60 C |
10 sec |
||||
| 65-95 |
10 sec |
1 |
|||
https://docs.google.com/spreadsheet/ccc?key=0An9mivRT-woWdGk1bWtjZG9SSTVBS2h1M1BwMURMSVE&usp=sharing
| Working label |
Primer Set |
Developmental Process |
|
| 3 |
Rp_RNF66_F |
Rp_RNF67_R |
multicellular organismal development |
| 4 |
Rp_flilCG1484_F |
Rp_flilCG1485_R |
muscle fiber development |
| 5 |
Mm_flilCG1484_F |
Mm_flilCG1485_R |
nematode larval development |
| 9 |
Mm_HCA57_F |
Mm_HCA57_R |
embryonic development ending in birth or egg hatching |
May 12, 2014
Gel electrophoresis of PCR products.
Used 12 ul sample per well -- 5 ul of HyperLadder II where indicated in table below
set to 120 V for 45 min
Gel Set up:
| Lane |
Primer |
| 1 |
Ladder (2000 bp) |
| 2 |
1 |
| 3 |
2 |
| 4 |
3 |
| 5 |
4 |
| 6 |
5 |
| 7 |
6 |
| 8 |
7 |
| 9 |
8 |
| 10 |
9 |
| 11 |
10 |
| 12 |
NTC |
| 13 |
|
| 14 |
1 |
| 15 |
2 |
| 16 |
3 |
| 17 |
4 |
| 18 |
5 |
| 19 |
6 |
| 20 |
7 |
| 21 |
8 |
| 22 |
9 |
| 23 |
10 |
| 24 |
NTC |
| 25 |
Ladder (2000 bp) |
| 26 |
|
| 27 |
|
| 28 |
| Working Label |
Primer Set |
Product Length |
Gene function |
|
| 1 |
Rp_dlgCG1725_F |
Rp_dlgCG1726_R |
162 |
determination of adult life span |
| 2 |
Rp_diaCG1768_F |
Rp_diaCG1769_R |
373 |
multicellular organismal development |
| 3 |
Rp_RNF66_F |
Rp_RNF67_R |
951 |
multicellular organismal development |
| 4 |
Rp_flilCG1484_F |
Rp_flilCG1485_R |
527 |
muscle fiber development |
| 5 |
Mm_flilCG1484_F |
Mm_flilCG1485_R |
319 |
nematode larval development |
| 6 |
Rp_skoCG3937a_F |
Rp_skoCG3937a_R |
332 |
nervous system development |
| 7 |
Rp_skoCG3937b_F |
Rp_skoCG3937b_R |
346 |
post-embryonic development |
| 8 |
Rp_ACVR_F |
Rp_ACVR_R |
874 |
sarcomere organization |
| 9 |
Mm_HCA57_F |
Mm_HCA57_R |
610 |
embryonic development ending in birth or egg hatching |
| 10 |
Rp_CSRP2BP_F |
Rp_CSRP2BP_R |
425 |
embryonic development ending in birth or egg hatching |
Results
Very bright positive for Primer 5-- Meretrix flilCG1484 (nematode larval development)looks like some contamination in negatve control-- but product is same length as well next to it (used primer 10 in neg control), most likely from loading the gel
Some of these are not the correct product length... Need to get a picture of the ladder key.
It looks like the bands from Meretrix are brighter-- but if two products or not the right length not sure what this really means.
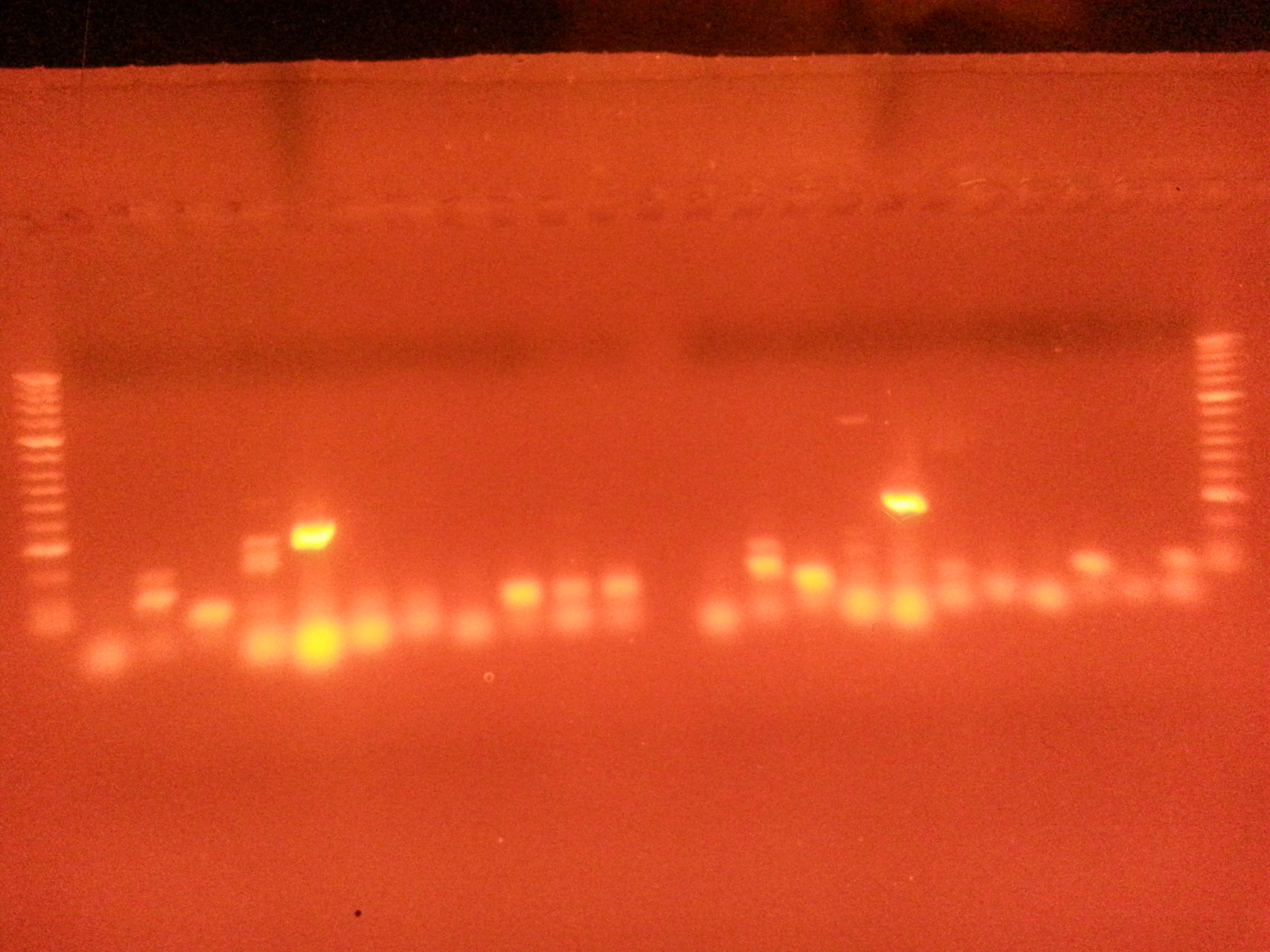
Notes and next steps:
really wish I had kept the gel a little longer to take a better pic. These are really blurry. Maybe make another
only need 100 V, 15 min. next time.
qPCR! check out melting curve make sure there's one product.
RNA extraction of larvae? if time allows..
May 9, 2014
Made 1.5% agarose gel-- 150 ml gel tray, 2.25 g agarose, 12 ul Ethidium Bromide.
28 wells-- will need 1000 bp ladder
most likely 15 ul in each well.
Stored in 209 fridge over weekend
May 8, 2014
Conventional PCR for all 10 primer sets in duplicate| stock concentration |
volume per reaction (ul) |
volume for cocktail |
|
Neg. Control |
Total |
Notes** |
|
| Water |
-- |
8.5 |
187 |
20 |
2 |
22 |
*5 primer sets in duplicate |
| Primer - F (uM) |
10 |
1 |
22 |
||||
| Primer - R (uM) |
10 |
1 |
22 |
||||
| GoTaq Green Master Mix (2X) |
1X |
12.5 |
275 |
||||
| 0 |
|||||||
| cDNA |
2 |
42 |
|||||
| Total volume (ul) |
25 |
550 |
|||||
| Thermalcycler conditions |
Used "Ernie" |
||||||
| Step |
Temperature |
Time |
Cycles |
||||
| Denaturation |
95C |
5min |
1 |
||||
| Denaturation |
95C |
30sec |
40 |
||||
| Annealing |
55C |
30sec |
|||||
| Extension |
722C |
90sec |
|||||
| Final Extension |
72C |
3min |
1 |
||||
| Hold |
4C |
∞ |
1 |
||||
| Working Label |
Primer Set |
|
| 1 |
Rp_dlgCG1725_F |
Rp_dlgCG1726_R |
| 2 |
Rp_diaCG1768_F |
Rp_diaCG1769_R |
| 3 |
Rp_RNF66_F |
Rp_RNF67_R |
| 4 |
Rp_flilCG1484_F |
Rp_flilCG1485_R |
| 5 |
Mm_flilCG1484_F |
Mm_flilCG1485_R |
| 6 |
Rp_skoCG3937a_F |
Rp_skoCG3937a_R |
| 7 |
Rp_skoCG3937b_F |
Rp_skoCG3937b_R |
| 8 |
Rp_ACVR_F |
Rp_ACVR_R |
| 9 |
Mm_HCA57_F |
Mm_HCA57_R |
| 10 |
Rp_CSRP2BP_F |
Rp_CSRP2BP_R |
Created working primer stocks-- dilution from 100 uM to 10 uM. stored in FTR 209 freezer
May 6, 2014
Goals:Conventional PCR to test three primer sets
Reverse transcription of more RNA
Only had enough cDNA for three primers in duplicate (6 wells)
http://replygif.net/t/facepalm
Three cocktail mixes-- one for each primer set.
Forgot a negative control --> will run these primer sets with others in next PCR.
http://replygif.net/t/facepalm (another)
| stock concentration |
volume per reaction (ul) |
volume for cocktail |
number of samples |
Notes |
|
| Water |
-- |
8.5 |
17 |
6 |
*3 primer sets in duplicate |
| Primer - F (uM) |
10 |
1 |
2 |
||
| Primer - R (uM) |
10 |
1 |
2 |
||
| GoTaq Green Master Mix (2X) |
1X |
12.5 |
25 |
||
| cDNA |
2 |
4 |
|||
| Total volume (ul) |
25 |
50 |
|||
| Thermalcycler conditions |
|||||
| Step |
Temperature |
Time |
Cycles |
||
| Denaturation |
95C |
5min |
1 |
||
| Denaturation |
95C |
30sec |
40 |
||
| Annealing |
55C |
30sec |
|||
| Extension |
722C |
90sec |
|||
| Final Extension |
72C |
3min |
1 |
||
| Hold |
4C |
∞ |
1 |
||
1. Rp_dlgCG1725
2. Mm_HCA57
3. Rp_skoCG3937b
Reverse transcription:
| Per Rxn |
Total used |
|
| RNA |
17.75 |
177.5 |
| oligo dT |
0.5 |
5 |
| 70C for 5 min |
||
| Store on Ice |
||
| Master Mix |
||
| M-MLV RT buffer (5X) |
5 |
50 |
| dNTPs (10mM) |
1.25 |
12.5 |
| M-MLV RT |
0.5 |
5 |
Primer Database:
https://eagle.fish.washington.edu:5001/fbsharing/z5naeMot
April 1, 2014
"Genomic resource development for shellfish of conservation concern"
http://onlinelibrary.wiley.com/doi/10.1111/1755-0998.12052/full
^ Link to Emma's paper as an example/resource of how to incorporate BLASTx into methods and materials
When citing BLASTx algorithm:
, , et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research, 25, 3389–3402.
To search for key terms within a column in SQLshare:
- "Derive Dataset"
- Enter source table name
- SELECT * FROM [jpb23@washington.edu].[mRNA taxid6580 joined_uniprot_sprot2]
- Command (without asterisks)
- where *insert column name* like '*insert target term*'
- Execute Query
March 11, 2014
Running 2 BLASTs on clam database taxonomic ID #65801. EST (Meretrix meretrix)
2. mRNA (Ruditapes phillippinarum)
http://nbviewer.ipython.org/url/eagle.fish.washington.edu/scaphapoda/Jessica/ipython_nb/Geoduck_hunt_EST.ipynb
http://nbviewer.ipython.org/url/eagle.fish.washington.edu/scaphapoda/Jessica/ipython_nb/Geoduck_hunt%20_Ruditapes.ipynb
Joined via SQLshare
https://sqlshare.escience.washington.edu/sqlshare/#s=home
to UniProt/Swiss-Prot database to annotate genes
March 10, 2014
To Do:
- Finish Methods and Materials to not lose Friday
- add qPCR, BLAST/primer choices, stats
- add company names
- add method of removing settlers
- change scientific name to generosa
- Remove settlers from sand
- measure geoducks and sand
- sieve from Friedman lab/Mac
- Isolate RNA from samples
- spin down all vials
- List of Capstone reagents with Charles
- 489 meeting
- 495 paperwork
- blog post
- keep up lit search
March 4, 2014
Reverse Transcription Protocol:A single reaction volume = 25uL.
- Aliquotted 17.75uL RNA to 0.5mL snap cap tubes.
- Add 0.5uL of Promega oligo dT for total volume of 18.25uL.
- Heat samples at 70C for 5 min in thermocycler.
- Place samples on ice IMMEDIATELY.
- Make Master Mix:
5 uL 5x Buffer (M-MLV RT Buffer)
1.25 uL 10mM dNTPs
0.5 uL M-MLV RT
*Processed one reaction. For future reactions perhaps double the volumes for each sample used (may be pipetting errors with small volumes)
7. Mix well.
8. Add 6.75 uL of master mix to RNA
9. Mix well, but do not vortex.
10.Incubate @ 42C for 1hr in thermalcycler
11.Heat inactivate @ 95C for 3 min.
12.Spot spin.
13.Store @ -20C.
qPCR :
| Volume |
Concentration |
||
| Sso Fast EvaGreen Supermix |
|||
| Primer F |
|||
| Primer R |
|||
| Water |
|||
| Template |
- Fill in table, add master mixes (X3), qPCR thermal conditions for SsoFast, and layout of wells
Primers in Boxes 11 and 12; Laternula ellipticca HSP and HSC
| 1561 |
Le_HSP70A_F |
CTGTCTTGAGCGATGGTGGC |
| 1560 |
Le_HSP70A_R |
TTTGTTACGGTCTTTCCTAAGTA |
| 1559 |
Le_HSP70B_F |
AAGCTTGTCAACCACGGCGG |
| 1558 |
Le_HSP70B_R |
CCTTGACCCTTTGGCCAAGG |
| 1557 |
Le_HSC70_F |
CAATGACAACACTCGCCCCA |
| 1556 |
Le_HSC70_R |
TGTTGACAGTCTTTCCGAGGTA |
February 27, 2014
Treating RNA stock with TURBO DNA-free-- treating 25 ul RNA stock to practice with procedure in case of error and for qPCR testing to assure no DNA.
5 x 50ul reactions, 5.052 ul RNA stock per reaction
- 5.052 ul RNA (5ug) + 5 ul DNA-free buffer + 0.5 ul DNase reagent + 41.443 ul water in 0.5 ml tube
- *note* this volume of water is incorrect for the reaction here-- should be 34 ul. Next round of DNA-free treatment needs to be the reaction volume (48 ul) minus the volume RNA and 0.1 vol DNase buffer, 0.1 vol inactivation reagent. (45-5.052-5-1)
- incubate for 30 min at 37 C (water bath)
- Add 0.5 ul DNase reagent
- incubate for 30 min at 37C
- Add 0.5 vol DNase Inactivation reagent (5 ul)
- Incubate RT for ~2 min
- Centrifuge 10,000 x g for 1.5 min
- Transfer to new tube-- combined tubes into one
Next steps:
- order primers
- qPCR of treated and untreated samples
- should see amplification in untreated, no amplification of treated.
- make cDNA out of DNase treated samples
- bake cookies for Taylor Shellfish :)
- Call Sarah about samples-- ask Brent about trips
February 23, 2014
Laternula elliptica --
| Primer Set |
Gene |
Primer Sequence |
RSq |
PCR Efficiency |
|
| 2F and 2Rev |
HSP70A |
Lel2F |
CTGTCTTGAGCGATGGTGGC |
0.998 |
116.80 |
| Lel2Rev |
TTTGTTACGGTCTTTCCTAAGTA |
||||
| 4F and 4Rev |
HSP70B |
Lel4F |
AAGCTTGTCAACCACGGCGG |
0.975 |
107.20 |
| Lel4Rev |
CCTTGACCCTTTGGCCAAGG |
||||
| 3F3 and 3Rev |
HSC70 |
Lel3F3 |
CAATGACAACACTCGCCCCA |
0.996 |
84.30 |
| Lel3Rev |
TGTTGACAGTCTTTCCGAGGTA |
95°C, 5 min; 35 cycles of 95°C, 20 s; 55°C, 20 s; and 72°C for 40 s elongation step of 72°C for 5 min
HSC = heat shock cognate
From Clark et al 2008 -- Antarctic marine mollusks do have HSP70 response
February 14, 2014
Geoduck capstoneRNA isolation
followed lab protocol on RNA isolation bench (will attach picture)
clipped off about half the siphon of one juvenile, remainder is still in RNALater
500 ul Tri Reagent
homogenize
500 ul Tri Reagent
-- took total of about 25 minutes
200 ul Chloroform
vortex
-- very cloudy/milky
centrifuged at 12000 g 15 min 4 deg C
aqueous in new tube
500 ul isopropanol
rest at room temperature about 8 min
centrifuged 12000 g 8 minutes 4 deg C
Nanodrop results
| lambda |
230 |
| Abs. |
10.884 |
| A-260 10 mm |
224.743 |
| A_280 10 mm |
13.819 |
| 260/280 |
1.79 |
| 260/230 |
2.27 |
| ng/ul |
989.7 |
need units
To Do:
Proposal
check in with Taylor (have trocophores at least!)
Order primers (sheet to Greg, give to Robin)
reverse transcribe to cDNA
-- when doing next RNA isolations, try new kit from Sam in parallel
Experimental set up
brainstormlarvae-- sent from Taylor at various stages of growth (easiest for them, most direct)
juveniles( all at same age, would be good subjects for environmental stressors)
adult-- diferent gonadal development stages, might be difficult to work with, limits tissue types can use (mostly siphon, nonlethal)
Set Up:
15 individuals at each development stage -- primarily interested in pre and post settlement mortality events and effects of metamorphosis
- -D-stage veliger-- including some towards end of veliger stage (16-35 days at 16 deg C, 47 days at 14 deg C)
- -initial metamorphosis
- -mid metamorphosis (week 2 or 3)
- -finishing metamorphosis (week 6-8)
- -post larval stage
- -Juvenile
- -maybe trocophore too (ask Brent about possibilties-- numbers needed to get RNA extract and availibility)
Analysis:
RNA extractionqPCR-- look for HSP70 at each stage
To Do (besides everything):
Email Brent with project plan to send to Mexico contact for primers
Order TriReagent (email sent to Greg, waiting to hear back)
Extract RNA from sample juvenile
Research primers -- NCBI
continue lit search
http://www.pac.dfo-mpo.gc.ca/science/species-especes/shellfish-coquillages/geopath/develop-eng.htm
http://protectourshoreline.org/DNR/ComprehensiveLitReview.pdf (Brent co-authored)
January 27th, 2014
Capstone; Geoduck Panopea abrupta
http://wdfw.wa.gov/fishing/shellfish/geoduck/
http://wdfw.wa.gov/fishing/commercial/geoduck/
http://wdfw.wa.gov/fishing/shellfish/geoduck/geoduck_references.html
References on Geoduck Clams (from wdfw.wa.gov)
- Bradbury, A. and J.V. Tagart. 2000. Modeling geoduck, Panopea abrupta (Conrad, 1849) population dynamics.II. Natural mortality and equilibrium yield. J. Shellfish Research, 19(1):63-70.
- Bradbury, A., B. Sizemore, D. Rothaus and M. Ulrich 2000. Stock assessment of subtidal geoduck clams (Panopea abrupta) in Washington. WDFW, Technical Report Number FPT00-01. 57 p.
- Goodwin, C.L.1976. Observations on spawning an growth of subtidal geoducks (Panope generosa, Gould). Proc. Nat. Shellfisheries Assoc. 65:49-58.
- Goodwin, L. and W. Shaul.1984. Age, recruitment and growth of the geoduck clam (Panope generosa, Gould) in Puget Sound, Washington. Wash. Dept. Fish. Progress Rpt. No. 215. 30 p.
- Goodwin, C.L. and B. Pease.1989. Species profiles: life histories and environmental requirements of coastal fishes and invertebrates (Pacific Northwest) -- Pacific geoduck clam. U.S. Fish. Wildl Serv. Biol. Rep. 82(11.120). US Army Corps of Engineers, TR EL-82-4. 14 p.
- Hoffmann, A., A. Bradbury and C.L. Goodwin.2000. Modeling geoduck , Panopea abrupta (Conrad, 1849) population dynamics. I. Growth. J. Shellfish Research, 19(1):57-62.
- Sizemore, B. and C. Blewett.2006. Geoduck studies in Hood Canal. Progress work associated with House Bill 1896. Report to the 2006 legislature, House Select Committee on Hood Canal. January 1, 2006. DNR, Aquatic Resources Division. 54 p.
- Washington Department of Natural Resources2000. The Puget Sound Commercial Geoduck Fishery Management Plan. December 2000. DNR, Aquatic Resources Division. 15p.
- Washington Department of Natural Resources.2001. Final Supplemental Environmental Impact Statement (FSEIS) for the State of Washington Commercial Geoduck Fishery. May 23, 2001.DNR, Aquatic Resources Division. 135 p.
Preliminary GenBank searches result in many Pab1-9, 12-19 etc cloned microsatellites, 18 S and 28S rRNA.
=
=
=
=
September 23rd 2013
Oyster HistologyTissue catalog completed with notation for questions and photo notes.
When in town, the last three weeks have been spent completing cuboidal metaplasia count for all new slides (8 slides of 1000 uatm, 8 slides of 1000 uatm + mech, 3 slides 400 uatm, 2 slides 400 uatm + mech, 6 slides 2800 uatm, 2 slides 2800 uatm + mech). Latest metaplasia and tissue data have been added to "Oyster Histology" Excel file on personal computer and uploaded to Eagle.
http://eagle.fish.washington.edu/scaphapoda/Jessica/Oyster%20Histology.xlsx
Metaplasia worksheet:
| Treatment |
Slide # |
Tubules open |
Total tubules |
Percentage open |
| 400 uatm |
H2.74* |
16 |
266 |
6.02% |
| 400 uatm |
H2.80 |
172 |
1,339 |
12.85% |
| 400 uatm |
H2.90 |
46 |
159 |
28.93% |
| 400 uatm |
H2.95 |
189 |
575 |
32.87% |
| 400 uatm |
H2.96 |
291 |
1,268 |
22.95% |
| 400 uatm |
H2.93 |
8 |
150 |
5.33% |
| 400 uatm |
H2.73 |
42 |
192 |
21.88% |
| 400 uatm |
H2.75 |
130 |
727 |
17.88% |
| 400 uatm |
H2.76 |
148 |
1257 |
11.77% |
| 400 uatm + mech |
H2.82* |
58 |
426 |
13.62% |
| 400 uatm + mech |
H2.84* |
20 |
203 |
9.85% |
| 400 uatm + mech |
H2.86* |
0 |
199 |
0.00% |
| 400 uatm + mech |
H2.87* |
15 |
324 |
4.63% |
| 400 uatm + mech |
H2.88* |
9 |
131 |
6.87% |
| 400 uatm + mech |
H2.85 |
63 |
654 |
9.63% |
| 400 uatm + mech |
H2.81 |
148 |
984 |
15.04% |
| 400 uatm + mech |
H2.83 |
100 |
663 |
15.08% |
| 1000 uatm |
H2.121 |
16 |
289 |
5.54% |
| 1000 uatm |
H2.122 |
54 |
923 |
5.85% |
| 1000 uatm |
H2.123 |
174 |
503 |
34.59% |
| 1000 uatm |
H2.124 |
29 |
610 |
4.75% |
| 1000 uatm |
H2.125 |
113 |
716 |
15.78% |
| 1000 uatm |
H2.126 |
142 |
378 |
37.57% |
| 1000 uatm |
H2.127 |
46 |
341 |
13.49% |
| 1000 uatm |
H2.137 |
42 |
886 |
4.74% |
| 1000 uatm + mech |
H2.129 |
0 |
834 |
0.00% |
| 1000 uatm + mech |
H2.130. |
68 |
799 |
8.51% |
| 1000 uatm + mech |
H1.131 |
124 |
1190 |
10.42% |
| 1000 uatm + mech |
H2.132 |
73 |
1007 |
7.25% |
| 1000 uatm + mech |
H2.133 |
169 |
741 |
22.81% |
| 1000 uatm + mech |
H2.134 |
0 |
451 |
0.00% |
| 1000 uatm + mech |
H2.135 |
155 |
1210 |
12.81% |
| 1000 uatm + mech |
H2.136 |
245 |
1337 |
18.32% |
| 2800 uatm |
H2.19 |
110 |
1,025 |
10.73% |
| 2800 uatm |
H2.21 |
0 |
613 |
0.00% |
| 2800 uatm |
H2.4 |
0 |
0 |
0.00% |
| 2800 uatm |
H2.5 |
140 |
707 |
19.80% |
| 2800 uatm |
H2.7 |
0 |
0 |
0.00% |
| 2800 uatm |
H2.17 |
140 |
686 |
20.41% |
| 2800 uatm |
H2.18 |
429 |
1016 |
42.22% |
| 2800 uatm |
H2.20 |
285 |
678 |
42.04% |
| 2800 uatm + mech |
H2.13 |
15 |
473 |
3.17% |
| 2800 uatm + mech |
H2.14 |
0 |
256 |
0.00% |
| 2800 uatm + mech |
H2.16 |
135 |
939 |
14.38% |
| 2800 uatm + mech |
H2.11 |
159 |
757 |
21.00% |
| 2800 uatm + mech |
H2.12 |
13 |
181 |
7.18% |
Photos of new slides:
| Name |
Position |
Objective |
Notes |
| H2.17.1 |
13 x 106 |
x10 |
|
| H2.17.2 |
13 x 106 |
x4 |
|
| H2.18.1 |
33 x 106 |
x10 |
|
| H2.18.2 |
34x105 |
x10 |
|
| H2.11.1 |
14 x 102 |
x4 |
|
| H2.12.1 |
18 x 104 |
x4 |
|
| H2.12.2 |
18 x 104 |
x10 |
possible example of dense hemocytes |
| H2.71.1 |
39 x 101 |
x10 |
comparative to normal mantle? |
| H2.83.1 |
16 x 105 |
x4 |
Intestine, sampling artifact |
| H2.83.2 |
20 x 107 |
x4 |
- currently on Friedman lab microscope, need to upload to Eagle from there. Under Desktop folder "Jessica"
Notes on Microscope:
- AWB= Auto White Balance; center the view on an area absent of tissue and select AWB at the top of the screen, this will make it so any color/tint disappear and color of staining more obvious.
- Microscope cover- not over the back black box, it might knock the dials and they would have to be reset.
- Don't need to turn on far left camera, it's for a different camera. Just need the driver under the monitor.
To Do:
- Run stats including new slides
- Box and whisker plot-- may need to do it by hand or after downloading SPSS
- Analyze "normal" without mechanical stress to get clear understanding of hemocyte accumulation and mucous cell accumulation.
=
=
August 23rd 2013
Oyster HistologyUpdated "Oyster Histology 1" Excel file on personal computer with the new slides data and rearranged it in an easier format to look at (finally figured out how!) and added "Metaplasia" worksheet so all data is on one document in 2 sheets. Adding stats data to worksheet next once finished with metaplasia data for new slides.
August 21st 2013
Oyster HistologySamples are in!
Preliminary cataloging for tissue types present, updating Excel file on SR lab desktop under Oyster Histology 1(1). Will upload new/cleaner file later on (after returning from work)New table should include latest samples with tissues present, which treatment groups all slides belong to, and additional notes. Cleaner version needs new layout-- present layout is very wide and not easily read.
As of right now it looks like only two slides do not include digestive gland. Unfortunately, both are from the 2800 uatm CO2 group, but at least we now have 6 slides to work with for that group instead of the original 2.
Ended day on counting dilated digestive gland lumen in slide H2.122-- took AutoCapture photos to start next round of data collection: AutoCapture 20102338p109--120.tif
August 14th 2013
Oyster HistologyBlog post up on monday the 12th following lab meeting
Selected and shipped off new samples to be stained
Slides kept in 70% ethanol, from Feb. 2012
Shipping Samples (in ethanol) for Central Histology Facility
(R. Strenge)- Wrap cassettes in paper towels and place in a ziplock bag.
- Add 30 ml of ethanol to the paper towels. It is OK to use multiple containers if there are many samples. (No more than 1 litre total per package).
- Place ziplockbag into another ziplock bag and place these in one of the small screw cap, plastic containers we purchased specifically for histology samples.
- Place the lid is on tight!
- Seal lid with 2 layers of parafilm.
- Place in a styrofoam shipping cooler. (This helps keep the samples cool and moist.)
- Add enough absorbent material (Sphagsorb or some from previous shipments) to equal the volume of ethanol being shipped.
- Add peanuts if necessary to keep container from moving around.
- Add instructions for CHF to cooler.
Inspected by Sam White, shipped with appropriate labels:

Slides Used
| Treatment Group (uatm) |
Slide # (H2.__) |
| 400 |
73, 74, 75, 76, 80, 90, 93, 95, 96 |
| 400 + mech |
81, 82, 83, 84, 85, 86, 87, 88 |
| 1000 |
121, 122, 123, 124, 125, 126, 127, 137 |
| 1000 + mech |
129, 130, 131, 132, 133, 134, 135, 136 |
| 2800 |
1, 3, 4, 5, 6, 7, 8, 17, 18, 19, 20, 21 |
| 2800 + mech |
9, 10, 11, 12, 13, 14, 15, 16, |
Treatment and corresponding sildes (all)
From Feb. 2012 sampling| Treatment Group (uatm) |
Slide # (H2.__) |
| 400 |
73, 74, 75, 76, 77, 78, 79, 80, 89, 90, 91, 92, 93, 94, 95, 96 |
| 400 + mech |
81, 82, 83, 84, 85, 86, 87, 88, |
| 1000 |
121-128, 137-138 |
| 1000 + mech |
129-136 |
| 2800 |
1, 3, 4, 5, 6, 7, 8, 17, 18, 19, 20, 21, 22, 23, 24 |
| 2800 + mech |
9, 10, 11, 12, 13, 14, 15, 16, |
=
=
August 10th 2013
Oyster HistologyMakeshift Box and Whisker plot of Control groups with and without stress... next is trying to make this more distinct, simple, and in an easier way.
1: without stress 2: with stress
When I tried adding axes labels the box portion went away..

Ran ANOVA test with single variable and repetition in Excel
One with high pCO2 running ANOVA between stressed and unstressed:
| Anova: Single Factor |
||||||
| SUMMARY |
||||||
| Groups |
Count |
Sum |
Average |
Variance |
||
| Hi pCO2 |
2 |
0.1073 |
0.05365 |
0.005757 |
||
| Hi pCO2 + stress |
3 |
0.1754 |
0.058467 |
0.0057 |
||
| ANOVA |
||||||
| Source of Variation |
SS |
df |
MS |
F |
P-value |
F crit |
| Between Groups |
2.78E-05 |
1 |
2.78E-05 |
0.004868 |
0.948765 |
10.12796 |
| Within Groups |
0.017156 |
3 |
0.005719 |
|||
| Total |
0.017184 |
4 |
||||
And another for the controls, stressed and unstressed:
| Anova: Single Factor |
||||||
| SUMMARY |
||||||
| Groups |
Count |
Sum |
Average |
Variance |
||
| Control |
6 |
1.0895 |
0.181583 |
0.01391 |
||
| Control + stress |
6 |
0.446 |
0.074333 |
0.002247 |
||
| ANOVA |
||||||
| Source of Variation |
SS |
df |
MS |
F |
P-value |
F crit |
| Between Groups |
0.034508 |
1 |
0.034508 |
4.271406 |
0.065642 |
4.964603 |
| Within Groups |
0.080788 |
10 |
0.008079 |
|||
| Total |
0.115295 |
11 |
August 7th 2013
Oyster Histology1:45-3:30
Met with Carolyn Friedman to go over slides and use of the microscope
- How do I distinguish between hemocytes and connective tissue nuclei?
- Hemocytes will be a dark purple nucleus surrounded by pink cytoplasm floating between the connective tissue
- Nuclei will be surrounded by the connective tissue/ embedded
- Accumulation of hemocytes is normal around intestine to a certain degree.
- sample of inflammation was left at the microscope
- Best way to get an idea of differences in accumulation could be to have an idea of "normal" then assign rankings as the degree of inflammation changes. ie. have "normal" be "0", a larger area or more dense accumulation be "1" and up from there.
- might be difficult to get quantitative data
- make sure using consistent objective lens.
- Is there an easier way to go through a slide? ie is there a way to mark a photo and keep that on a fixed location? Other tips and tricks?
- not really..... just keep using the Auto Capture and be very aware of the borders
- But to pinpoint a location on a slide, write down the coordinates using the scales on 2 sides of the slide
- The black bar on the bottom of the screen on NIS Elements-- the number goes up when more in focus, you know you're moving out of focus again when the number starts going down
- The flat bar under the stage-- when on 4x push it in, when above 4x pull it out.
- Make sure my idea of metaplasia/ dilated lumen is correct
- looks good! yay!
- Insert snapshot of cuboidal metaplasia
- Called cuboidal because the columnar cells that make up the lumen turn more cube-shaped
- Chloride cells? Viable option?
- Since oysters are osmoconformers probably not
- Suggestions for the next moves?
- Just keep looking at the ones I know to be control without secondary stress to get a better idea of what "normal" looks like
- Broad-scale view of hemocytes to see if obvious accumulation
- Mucous cells? (Still not sure how to approach)
- Fluorescent antibody test-- HSP 70
- heat shock protein that usually is expressed when stressed
- Went over a few slides with unknown tissue types
- kidney guess was correct, other unknown tissue types included cross sections of epithelial tissue as well as oracle section of heart
- H2.90 Has section of kidney with dark pigment: ferratin (not sure if spelling is correct)
- Maybe check out which other slides have kidney to look at in future
- Hemocyte accumulation can occur around germinal follicles if they are being reabsorbed
Other things to look into:
How long after being spun were the oysters sampled?
How were they sampled? -- how long were they out of the water, were some out of the water longer than others etc.
What about using "JMP" instead of Excel? -- Check out what kind of learning curve that would be
To Do:
Blog Post
Look into hemocyte accumulation
Review control slides
Make Box and Whisker plot
Get R code from Emma?
August 2nd 2013
Oyster Histology10:00-11:30, 3:00- 4:30
Met with Emma
- start running stats
- found out which are control and which are treatment samples
| Treatment |
Slide # |
Tubules open |
Total tubules |
Percentage open |
| Control |
H2.74* |
16 |
266 |
6.02% |
| Control |
H2.80 |
172 |
1,339 |
12.85% |
| Control |
H2.90 |
46 |
159 |
28.93% |
| Control |
H2.95 |
189 |
575 |
32.87% |
| Control |
H2.96 |
291 |
1,268 |
22.95% |
| Control |
H2.93 |
8 |
150 |
5.33% |
| Control + stress |
H2.82* |
58 |
426 |
13.62% |
| Control + stress |
H2.84* |
20 |
203 |
9.85% |
| Control + stress |
H2.86* |
0 |
199 |
0.00% |
| Control + stress |
H2.87* |
15 |
324 |
4.63% |
| Control + stress |
H2.88* |
9 |
131 |
6.87% |
| Control + stress |
H2.85 |
63 |
654 |
9.63% |
| Hi pCO2 |
H2.19 |
110 |
1,025 |
10.73% |
| Hi pCO2 |
H2.21 |
0 |
613 |
0.00% |
| Hi pCO2 + stress |
H2.13 |
15 |
473 |
3.17% |
| Hi pCO2 + stress |
H2.14 |
0 |
256 |
0.00% |
| Hi pCO2 + stress |
H2.16 |
135 |
939 |
14.37% |
Problems:
- Excel doesn't run ANOVA with unbalanced sample sets (Control are 6-6, Treatment are 3-2)
- Very small samples
- No straightforward way to make box and whisker plot?
To Do:
- Make box and whisker plot
- Research how to run stats in Excel
- run stats through R
- Look into JMP
July 31st 2013
Oyster Histologyhttp://ac.els-cdn.com/S0022201196946261/1-s2.0-S0022201196946261-main.pdf?_tid=2791f4aa-fa05-11e2-810d-00000aab0f26&acdnat=1375291262_d7402c4609fc3471ee57bf9e85df59ea
July 30th 2013
Oyster Histology3:15-6:30 PM
Completed Digestive Tubule Count!
Both open and total with percentage to make sure it's not just a sampling error (some slides had more of the digestive gland included than others)| Slide # |
Tubules open |
Total tubules (7/24/13) |
Percentage open |
| H2.13 |
15 |
473 |
3.17% |
| H2.14 |
0 |
256 |
0.00% |
| H2.16 |
135 |
939 |
14.37% |
| H2.19 |
110 |
1,025 |
10.73% |
| H2.21 |
0 |
613 |
0.00% |
| H2.74* |
16 |
266 |
6.02% |
| H2.80 |
172 |
1,339 |
12.85% |
| H2.82* |
58 |
426 |
13.62% |
| H2.84* |
20 |
203 |
9.85% |
| H2.86* |
0 |
199 |
0.00% |
| H2.87* |
15 |
324 |
4.63% |
| H2.88* |
9 |
131 |
6.87% |
| H2.90 |
46 |
159 |
28.93% |
| H2.95 |
189 |
575 |
32.87% |
| H2.96 |
291 |
1,268 |
22.95% |
| H2.85 |
63 |
654 |
9.63% |
| H2.93 |
8 |
150 |
5.33% |
Other Notes/ What's next:
Heard back from staining company-- should be able to see chloride cells in gill tissue using current stains. Check this with Dr. Friedman and review the paper.Hemocyte accumulation: try using Image J to see if it will distinguish hemocytes (and chloride cells?) -- review Roberts paper
Run some stats! Let's see if any of this data is significant
- meeting with Emma on Wednesday to review how
July 29th 2013
Oyster Histology10:00-11:30 AM
Lab meeting--
Next unseminar 8/22?
Research symposium 8/26 for lil interns
need another blog post!
Image J tutorial wiki -- might start using Image J for hemocyte accumulation. Need to become a little more familiar with it first though.
http://imagejdocu.tudor.lu/doku.php?id=howto:start
July 26th 2013
Oyster Histology10:30-12:30 AM
Recounted degestive tubules expressing metaplasia
- testing methods
For slides H2.16 and H2.19 I used the count tool while keeping the camera feed live to re-count metaplastic cells
For slides H2.21-H2.80 I used the live feed to mark cells in a single screen, right click to total and to turn the markers red (rather than blue). Then I went through the same screen to count open lumen (while the markers were blue) and totaled that count. I then moved through the slides in rows doing my best not to overlap the photos.
Completed slides H2.16-H2.74. H2.80 had many more digestive tubules than the previous slides; I took pictures using Auto Capture and used the count tool for both total and open tubules.
I did not have time to complete slide H2.80 before having to go to work, ended on photo 20102338po32 (last one in autocapture folder)
To Do:
Finish slide H2.80- pics 26-35
Count digestive cells on remaining slides
Count open lumen on remaining slides
Calculate percentages
Review running some stats
Meet with Dr. Friedman about answering scope questions
Play with Image J for hemocyte accumulation
Review staining article-- Find out if histology company can stain for chloride cells
Blog post
Digestive tubule count updated to below table from July 18th
July 24th 2013
Oyster Histology3:45-6:00 PM
Counting Digestive cells
Methods:
using Friedman microscope to take pictures of all digestive tubules, then use count tool to mark each tubule.
Later used "AutoCapture" tool for easier access to pictures once taken. After using capture initially, NIS Elements became unresponsive. I reopened it and used autocapture and it worked without crashing again.
Problems:
- it's very difficult to tell where the borders between pictures are. I'm nervous I'm counting cells between pictures twice and can't tell where each pic starts and stop. (lots of just user error)
- I might need to go through and recount those expressing metaplasia. Looking at the slides a little closer I feel like there were more that I would consider having open lumen.
Questions:
- What is ROI? it came up when using autocapture
- Is there a way to get a picture of the entire slide? Or at least what is seen through the scope? As of right now the screen is only a small, zoomed-in portion of the bino.
- Is there a way to draw a shape, then move to another area while keeping the drawn image on the object in question? Ie. use the count tool to mark each tubule while moving around the entire slide?
- Is this even necessary? I feel like it would be good to get an idea of how much of a sampling error the metaplasia count could be but I want to make sure the info I'm getting here is useful
Play with Image J- for hemocyte count and for metaplasia
Run stats on metaplasia percentages
Review Roberts paper for hemocyte accumulation
Totals added to below table.
July 18th 2013
Oyster Histology9:45-10:30 AM; 3:30- 5:30 PM
Metaplasia search
What I was calling "possible metaplasia" yesterday is most likely a slice of intestine (now corrected). So, today the goal was to go through each slide looking specifically for open digestive gland lumen as an indicator of metaplasia.Notes: amount of digestive gland present varies pretty drastically between slides (leading to sampling errors) maybe use as percentage of total tubules present; requires count tool on Friedman microscope. See if there's a way to count two at once, to differentiate those "open" while counting total.
Intestine tissue tends to have more mucous cells and defined epithelial cells-- looks almost striated (use to differentiate between the two)
| Slide # |
Tubules open |
Total tubules (7/24/13) |
Percentage open |
| H2.13 |
15 |
473 |
3.17% |
| H2.14 |
0 |
256 |
0.00% |
| H2.16 |
135 |
939 |
14.37% |
| H2.19 |
110 |
1025 |
10.73% |
| H2.21 |
0 |
613 |
0.00% |
| H2.74* |
16 |
266 |
6.02% |
| H2.80 |
172 |
1339 |
12.85% |
| H2.82* |
58 |
426 |
13.62% |
| H2.84* |
20 |
203 |
9.85% |
| H2.86* |
0 |
199 |
0.00% |
| H2.87* |
15 |
324 |
4.63% |
| H2.88* |
9 |
131 |
6.87% |
| H2.90 |
46 |
159 |
28.93% |
| H2.95 |
189 |
575 |
32.87% |
| H2.96 |
291 |
1268 |
22.95% |
| H2.85 |
63 |
654 |
9.63% |
| H2.93 |
8 |
150 |
5.33% |
Open tubule count may change soon using the Friedman microscope.
Other miscellaneous tasks done today:
Fixed picture upload error from Eagle to wikispace; no more "#" in folder titles. Re-embedded photos from Eagle to July 17th table.
Conferred with Emma and Lisa about tissue types seen in July 17th table and whether or not metaplasia was present. Modified below table to reflect corrections (ie. identifying heart and kidney tissues, confirming germinal follicles)
To do:
Prepare slides for staining on Monday
- keep looking up literature on chloride cell staining and gill histology
Review stats notes/book to run preliminary stats on metaplasia
- see if can identify which treatment and which control
=
=
July 17th 2013
Oyster Histology2:30-5:00 PM
Exploring stress response detection methods
Goals:
- Test hemocyte counting methods for efficiency and for best representation
- for full screen vs given area circle
- Look again for signs of metaplasia
- be aware of malformed digestive tubules/ open tubules
- keep track of how many slides and which
- Look for mucous cells, see where accumulate most
- Find patterns of how tissues are arranged to see where focus loci
| Picture # |
Picture |
Notes |
| H2.13.1 |
 |
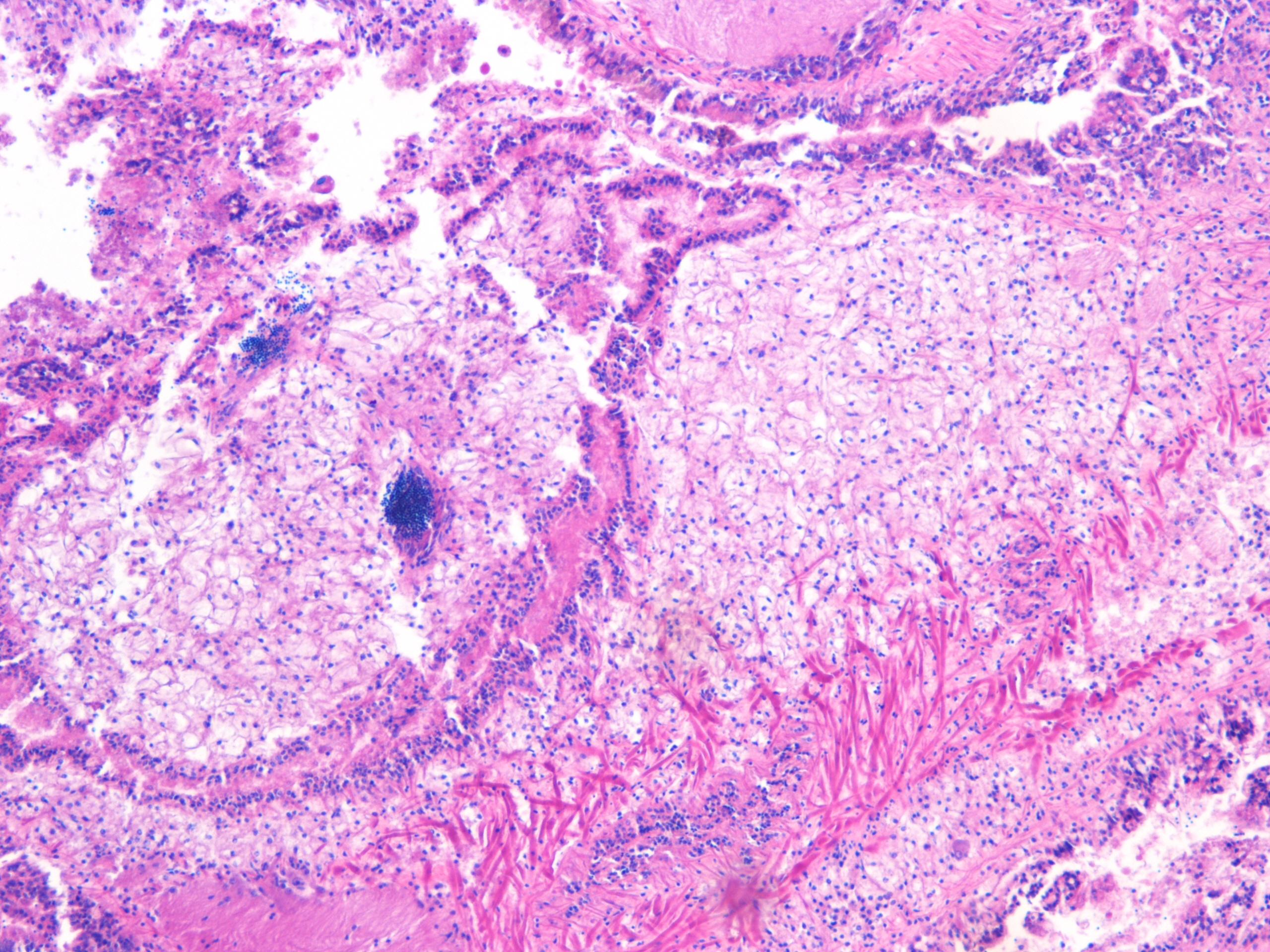
Possible metaplasia slices of intestine |
| H2.13.2 |
 |
A few open lumen-- metaplasia? center long tissue probably intestine.. |
| H2.13.3 |
 |
Germinal follicles |
| H2.13.4 |
 |
Unknown tissue |
| H2.13.5 |
 |
Closer view of unknown |
| H2.13.6 |
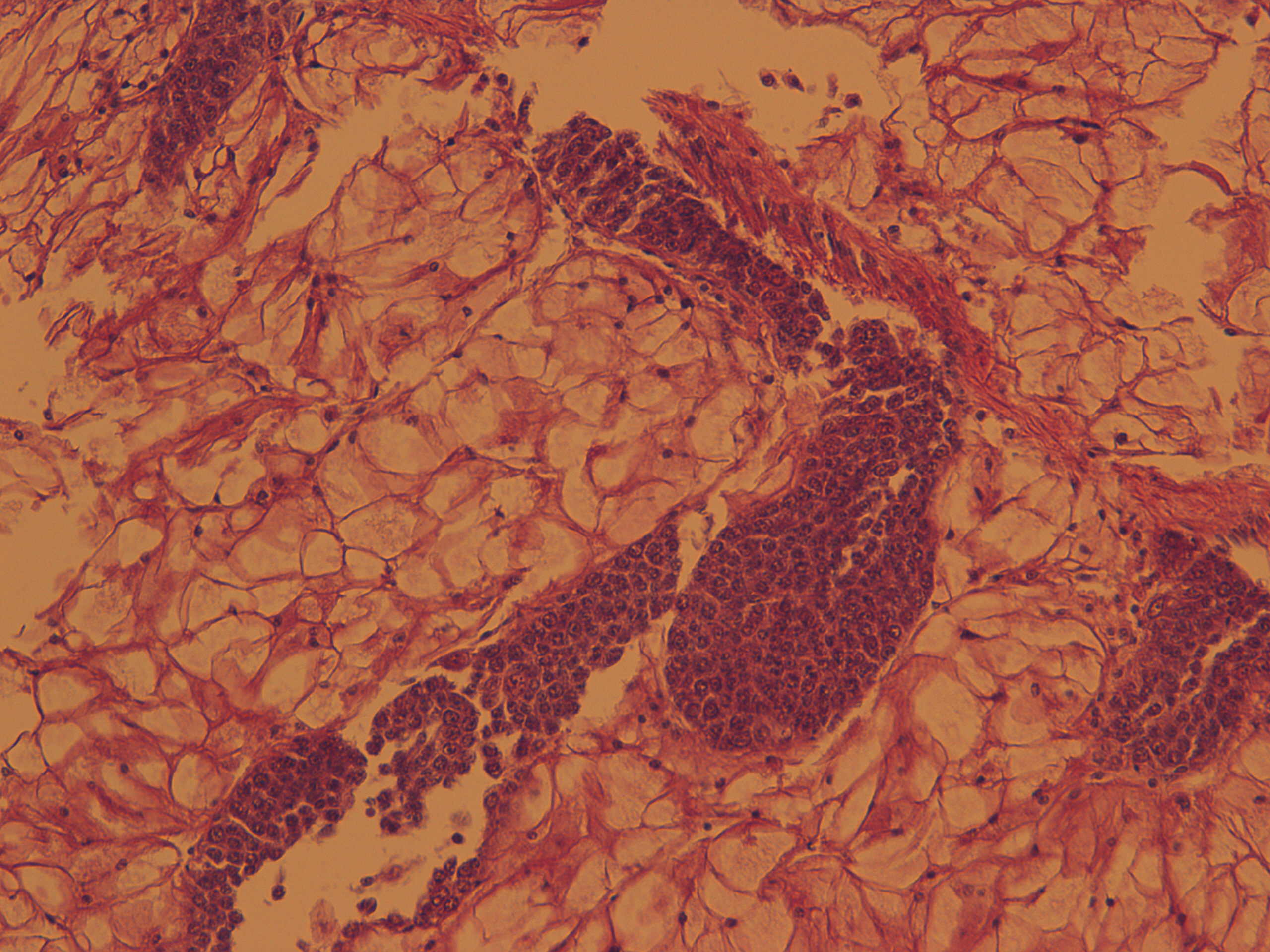
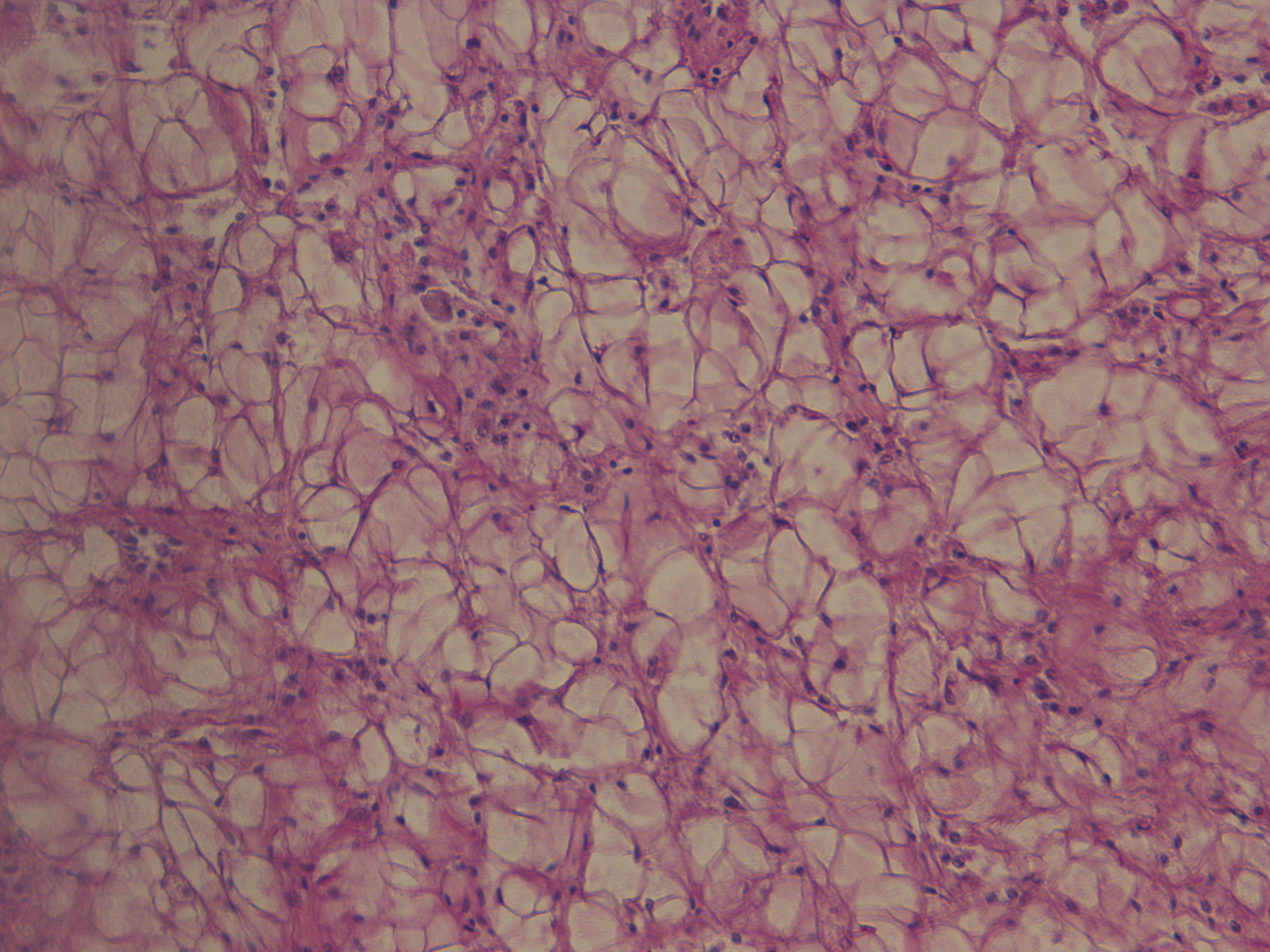 |
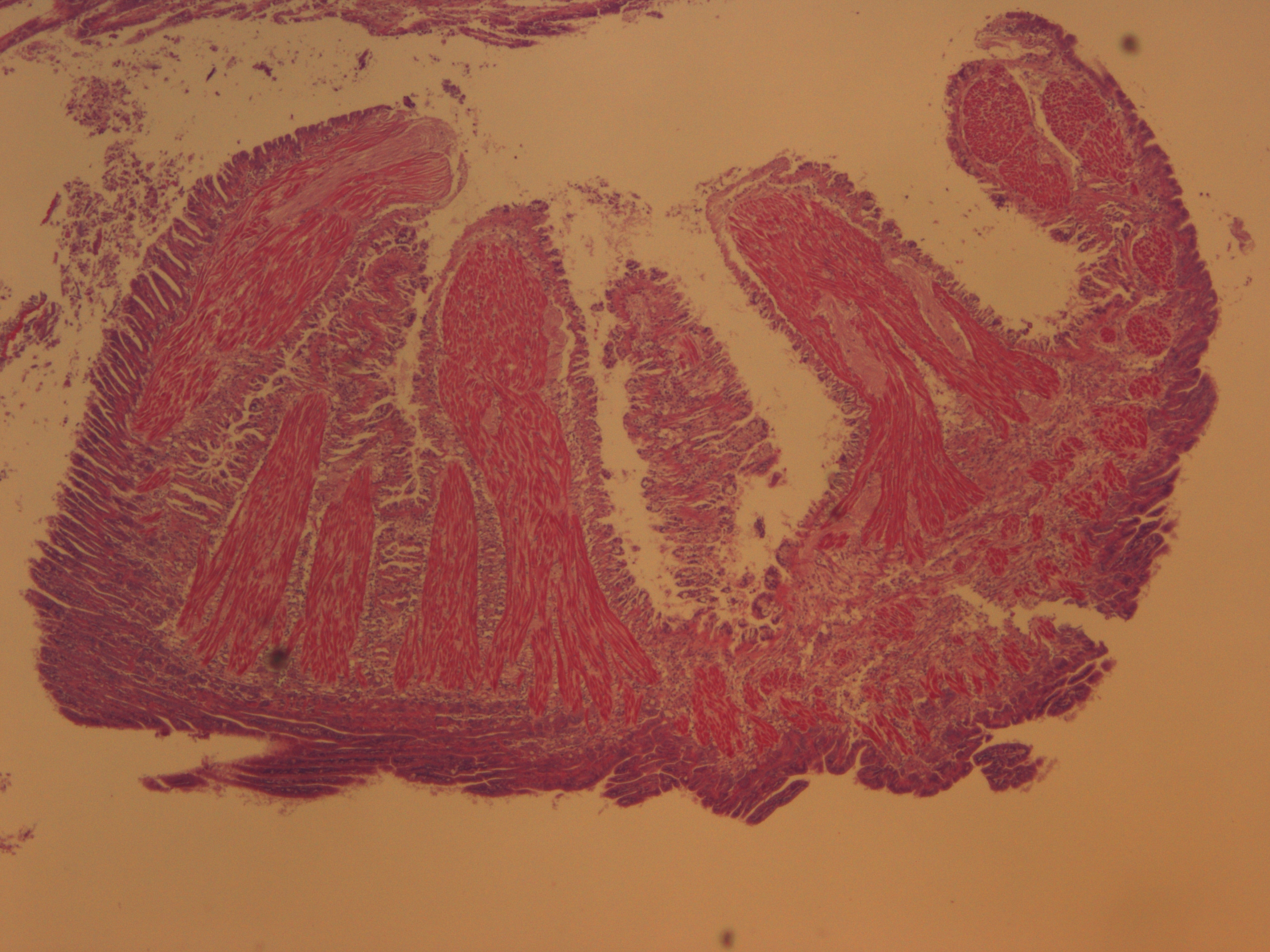
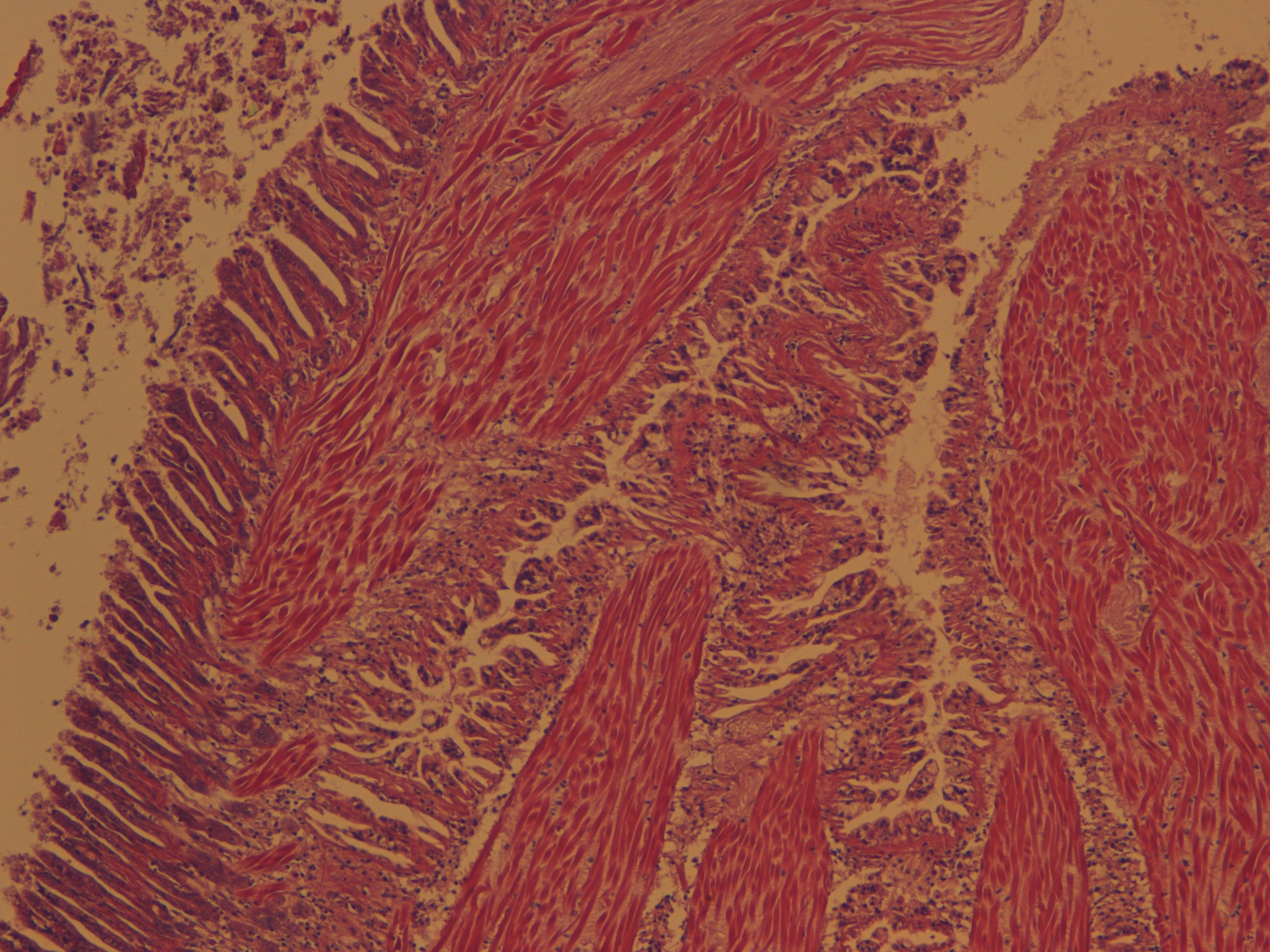
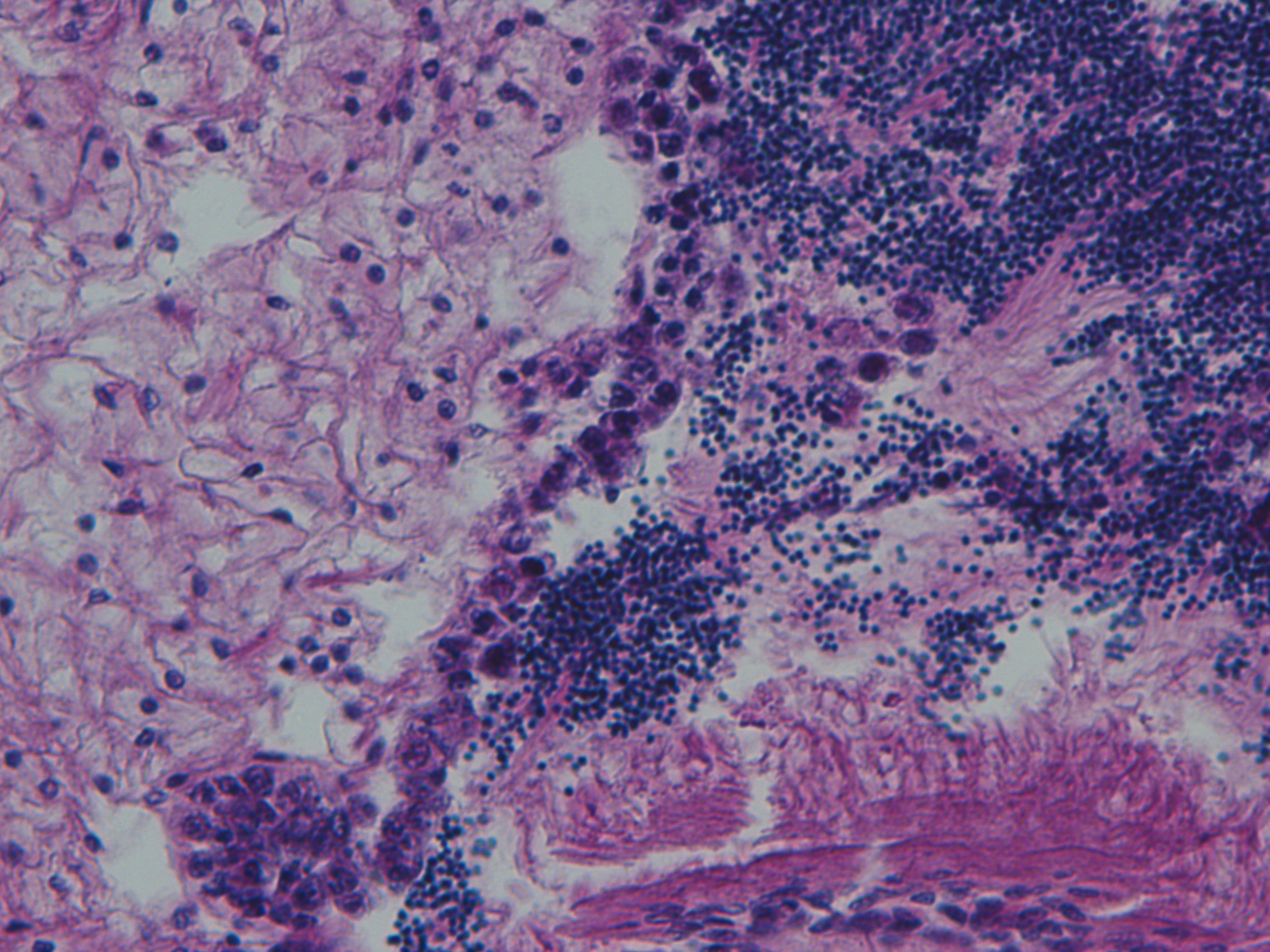
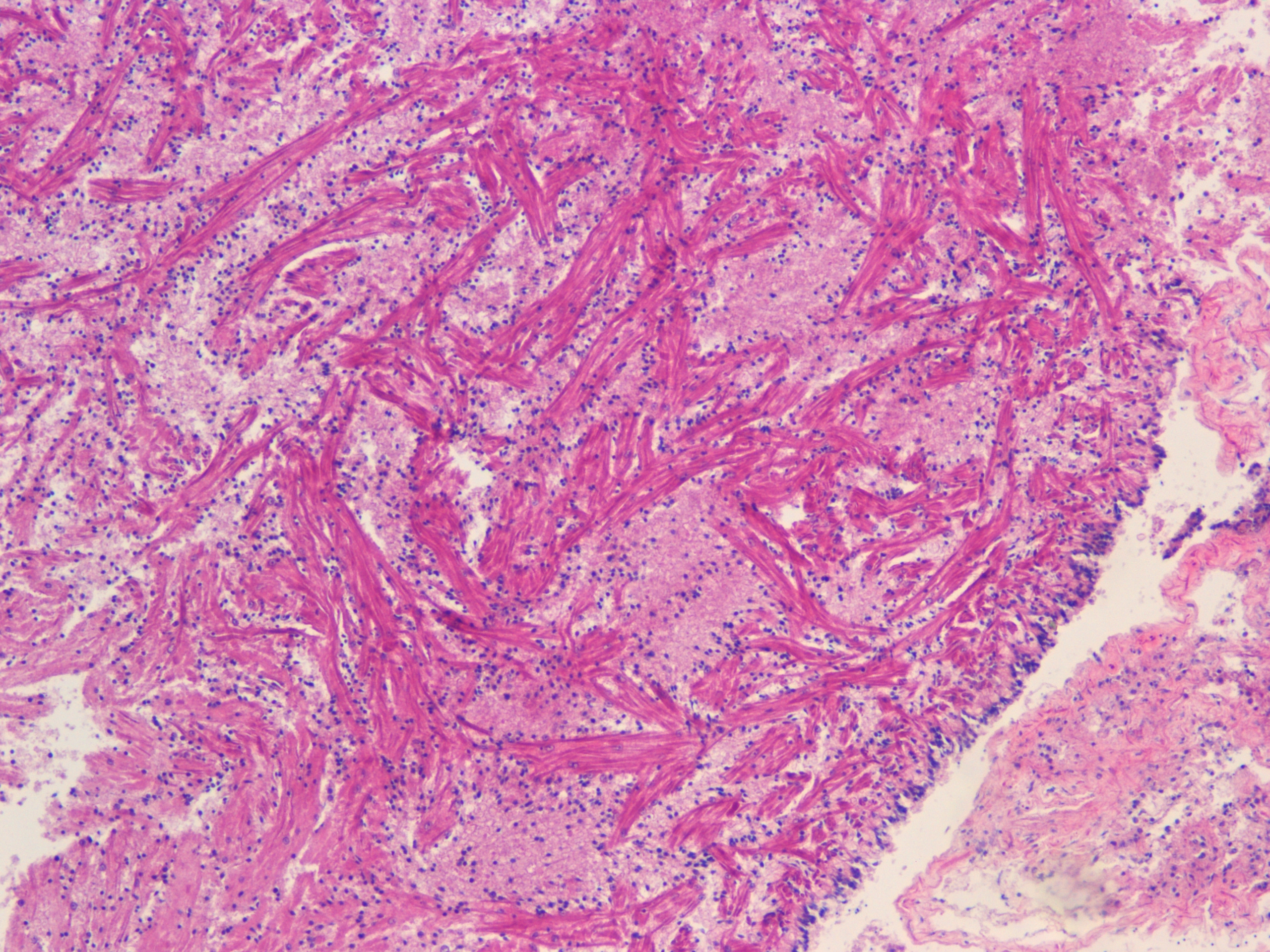
Test of hemocyte counting: Took area of mantle between digestive gland and intestine zoomed in to 20x (546.6px X 1913.3 px) used "count" tool to click on each hemocyte counted 505 hemocytes... It's possible that what I was counting may or may not be hemocytes (more beneficial to do smaller or larger area?) |
| H2.14.1 |
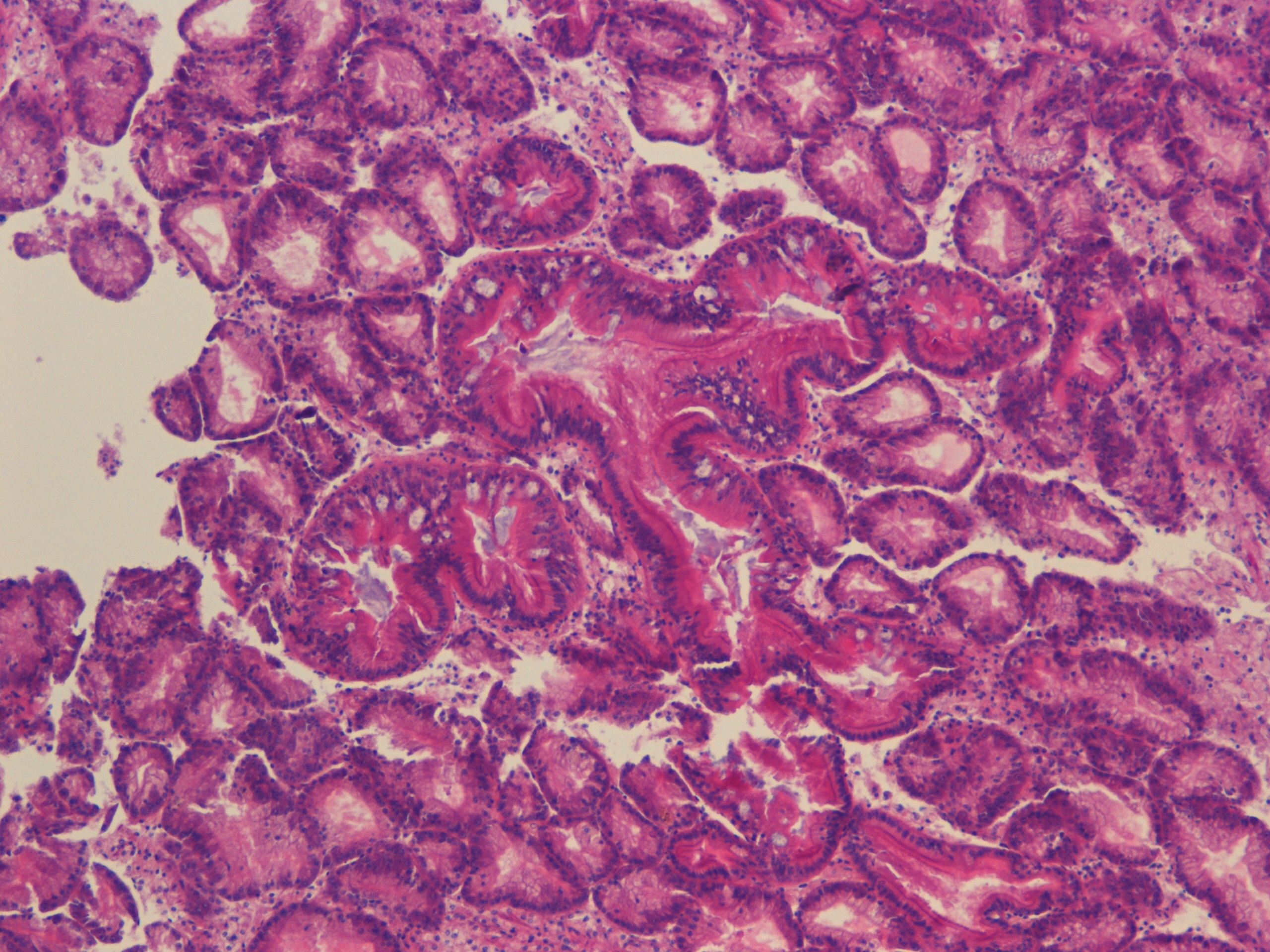 |
possibly metaplasia -- far left open lumen center of photo most likely slice of intestine |
| H2.14.2 |
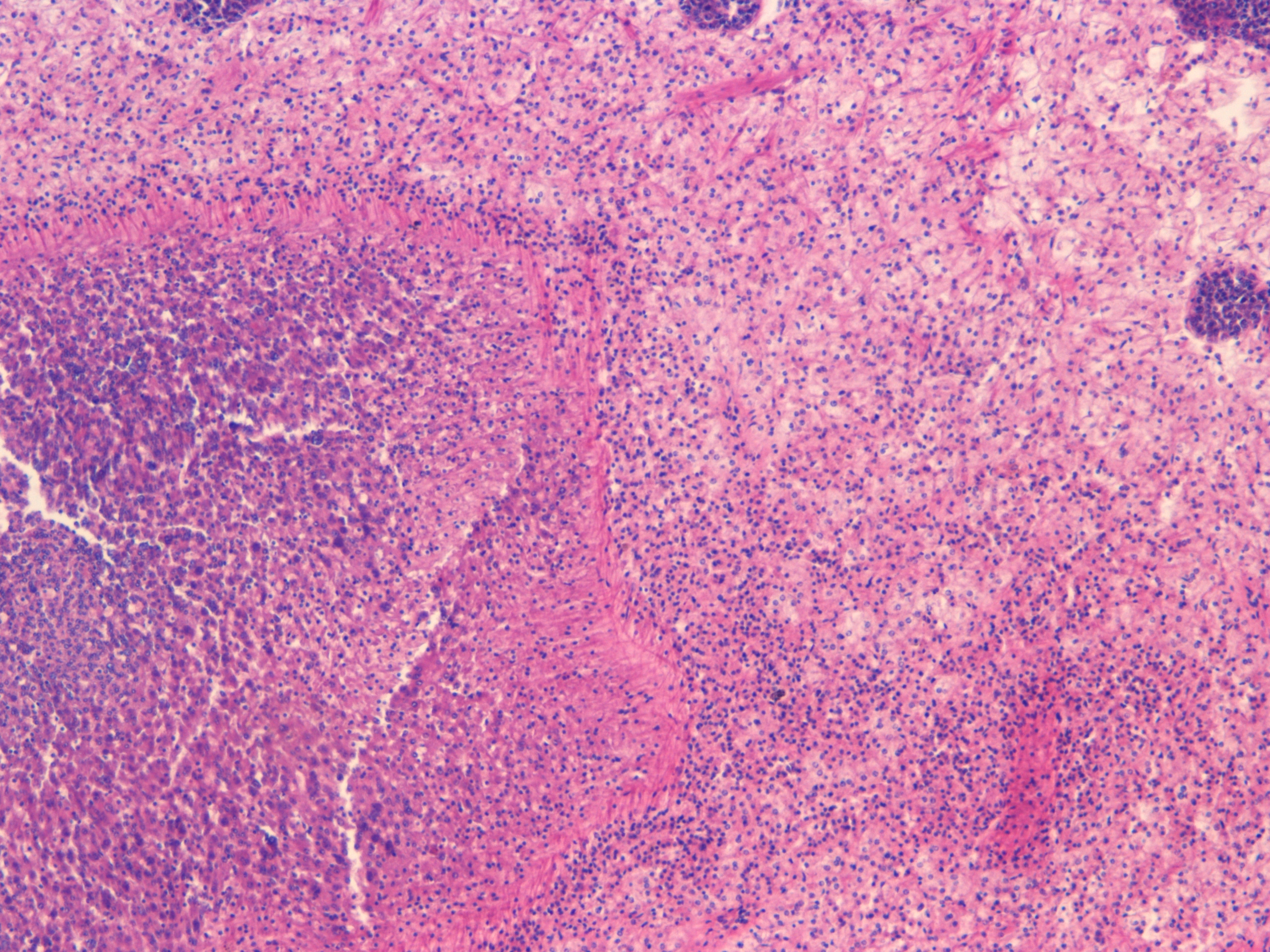 |
left side might be different tissue looks like accumulation of hemocytes but surrounded by some other tissue as well.. |
| H2.14.3 |
 |
possibly spermatids (better pic than "unknown" table) |
| H2.14.4 |
 |
Mantle, with reproductive follicles |
| H2.14.5 |
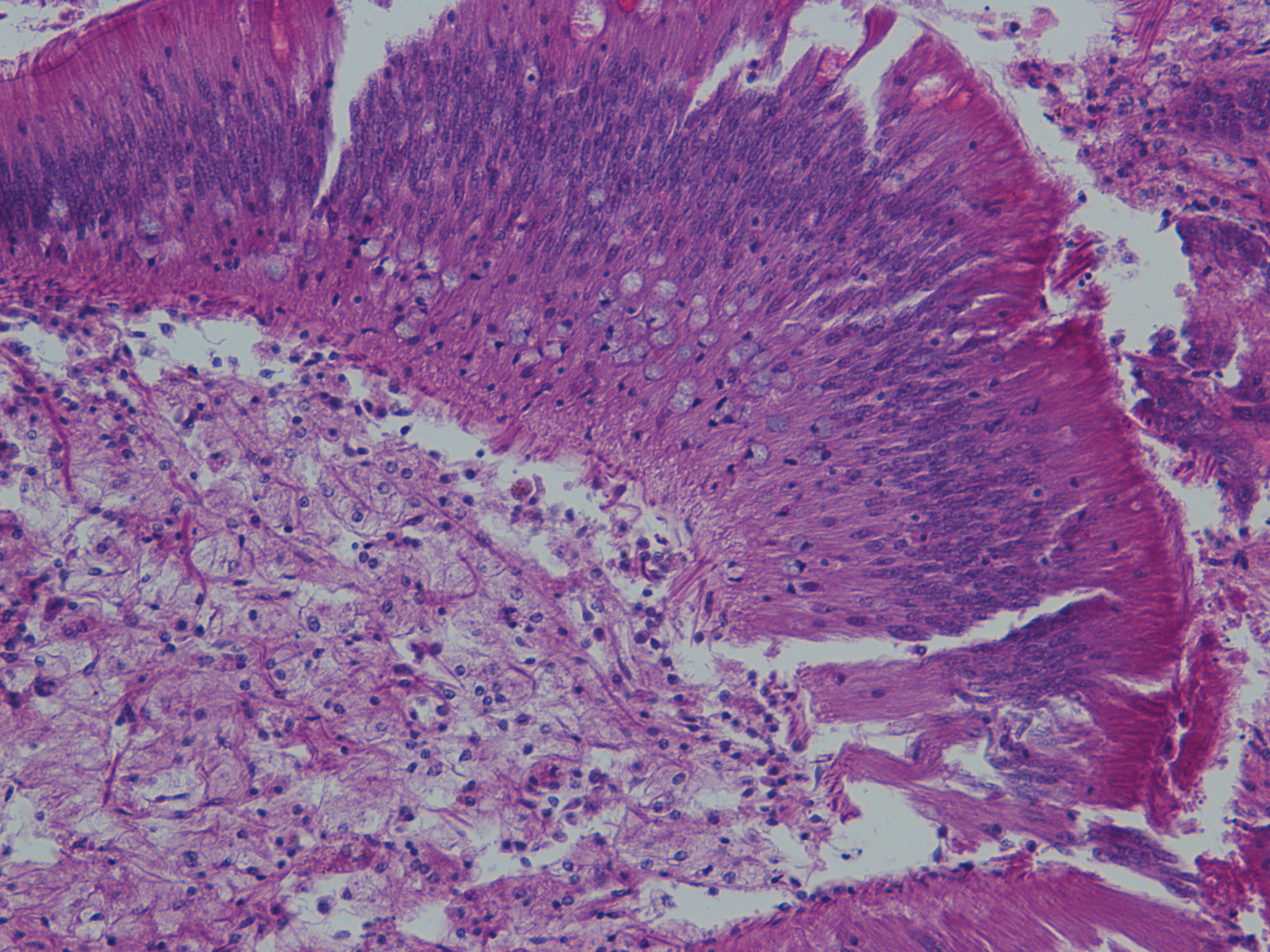 |
Intestine (shot of mucous cells) |
| H2.14.6 |
 |
Heart |
| Overall |
good examples of mucous cells! |
Notes:
Captured photos saved under Desktop folder = "Jessica"
Ran into some issues with the slide H2.14.2 picture upload-- continue later
All photos are uploaded to Eagle (C.gigas histology photos #2)
http://eagle.fish.washington.edu/scaphapoda/index.php?dir=Jessica%2FC.+gigas+histology+photos+%232%2F
- start bringing thumbdrive to Friedman lab or just upload all pics to Eagle as soon as finished with day.
Miscellaneous tasks done:
E-mailed Dr. Friedman
Blog post
Re-ordered slides-- far right is the start of the slides containing mantle, intestine, digestive gland, AND gill.
H2.13-H2.96
July 15th 2013
Oyster Histology10:00-11:20; 3:30-4:45
Lab meeting notes: up the blog posts, keep an eye out on the Google + community for lab meeting themes?
Slides containing mantle, gill, digestive gland, and intestine:
Representative photos and unknown tissue photos
Slide number listed on the left (if no slide number than it's a continuation of the previous slide)It's likely that these are the slides to be used in tissue analysis given the consistency of tissue type
Corresponding notes are in the Excel file below
http://eagle.fish.washington.edu/scaphapoda/index.php?dir=Jessica%2F
| H2.13 |
|||
| H2.14 |
|||
| H2.16 |
|||
| H2.19 |
|||
| H2.21 |
|||
| H2.74 |
|||
| H2.80 |
|||
| H2.82 |
|||
| H2.84 |
|||
| H2.86 |
|||
| H2.87 |
|||
| H2.88 |
|||
| H2.90 |
|||
| H2.95 |
|||
| H2.96 |
|||
Next:
Draft e-mail to Dr. Friedman about unknown tissues and how to proceed.
Inflammation analysis:
- Use microscope in Friedman lab to analyze hemocyte accumulation
- take a given area within target tissues and count hemocytes
- To do this, though, I need to be sure I have a clear idea of what hemocytes look like as well as the four tissue types to make sure I'm not calling it "inflamed" but it's actually another type of tissue
- Keep an eye out for mucous cells (look for discrepancies in size, number, and location in tissues)
Restain for chloride cells around gills
- buffered cosin and Harris' hematoxylin
July 10th 2013
Oyster Histology2:00-5:00 PM
It's official: my computer is too slow to embed pictures. Will be doing so from lab computers from now on...
Embedded pictures into below table
Figured out hot to post to the tumblr, next is OA blog
Reviewed Friedman binder to compare to unknown tissues and analyze metaplasia
- Morphological and morphometrical changes in chloride cells of the gills of Pimephales promelas after chronic exposure to acid water
- Stained in buffered cosin and Harris' hematoxylin
- http://link.springer.com/article/10.1007/BF00216521#page-1
- Results are cut off :/ maybe via UW libraries log in.
(Sidenote/ tangent thought: wondering how histology of Ulcerative Colitis would relate to acidic environments and act as an example of what to look for in inflamed intestine cells; primarily bacteria caused but ulcers usually related to acidic environment... how similar? http://www.sciencedirect.com/science/article/pii/S1542356513004606 <-- markers)
- Effect of Pollution History on Immunological Responses and Organ Histology in the Marine Mussel Mytilus edulis Exposed to Cadmium
July 8th 2013
Oyster Histology3:00-7:30 PM
Tutorial on how to use Friedman microscope with Emma and Claire.
- Turn on computer, monitor, camera , and driver
- NO SEAWATER
- button directly to the right of lenses controls whether bino or photo
- light adjustment on left, flat black knob
- NIS Elements
- Change Objective used in the bottom right corner of screen-- labeled "Nosepiece"
- remember to press play if not getting live feed, Stop to take pic (top bar with play button)
- Top catergories (ie File, Tools, etc) includes "Measure"; use this to get lengths, area etc
- Clear screen button above Objective
- Make sure to turn everything OFF when done. And turn objective to 4x.
Double check How To section to embed pictures from Eagle
Updated Excel file to sort comparable data and isolate unknown tissue types (not sure if this will translate into the previous link)
- 1 = has tissue
- 0 = tissue absent
- yellow highlight = contains all 4 tissues
Unknown Tissue Pictures:
Slide number on the far left, tissue sample will be on the right, most likely one pic at 4x, another 10x.Red text indicates that particular slide doesn't contain all four tissues (gill, intestine, digestive gland, mantle)-- so it's likely not to be included in comparative analysis
| H2.14 |
||||
| H2.16 |
||||
| H2.19 |
||||
| H2.74 |
||||
| H2.88 |
||||
| H2.90 |
||||
| H2.95 |
||||
| H2.10 |
||||
| H2.9 |
<-- bottom and top tissues surrounding the mantle; just mantle epithelium? |
|||
| H2.93 |
||||
| H2.93 (con) |
||||
July 7th 2013
Oyster HistologyContinue literature search:
Acute stress alters intestinal function of rainbow trout (Olsen et al)
http://www.sciencedirect.com.offcampus.lib.washington.edu/science/article/pii/S004484860500178X
-- food deprivation as stress but looked at different biochemical markers of stress (haemacrit, blood glucose, lactate, cortisol) and changes in permeability of mannitol
-- electron microscopy of enterocytes at different sections (hindgut, midgut etc)
Tissue Adaptation to physical stress (Mueller and Maluf)
http://web.ebscohost.com.offcampus.lib.washington.edu/ehost/detail?sid=9aa32740-1ecf-4c3b-8cd9-406ad0b982fb%40sessionmgr114&vid=1&hid=124&bdata=JnNpdGU9ZWhvc3QtbGl2ZQ%3d%3d#db=a9h&AN=6406638
This will probably be a good resource:
"Histological techniques for marine bivalve mollusks" (Howard and Smith)
http://permanent.access.gpo.gov/gpo7399/tmfnec25.pdf
July 3rd 2013
Oyster Histology10:00-11:30 AM; 2:00-5:00PM
Uploaded Excel file and histology slide photos to Eagle server--> Scaphopoda--> Jessica
http://128.95.149.81:5000/webman/index.cgi
http://eagle.fish.washington.edu/scaphapoda/index.php?dir=Jessica%2F
(renamed photo files, struggled with iPhone connections, followed Roberts lab on tumblr and Facebook)
Continued literature search...
- Histological and histochemical development of the digestive system of Solea senegalensis (Kaup, 1858) larvae (L. Ribeiro)
- http://www.sciencedirect.com/science/article/pii/S0044848698004967
- full: http://www.sciencedirect.com.offcampus.lib.washington.edu/science/article/pii/S0044848698004967
- Protien, pancreas, liver, gastric glands, esopphageal mucous
- Histology, histochemistry and enzyme biochemistry in the digestive system of the endosymbiont-bearing bivalve Loripes lucinalis
- Histological survey of Pacific Oysters in Galicia (D. Iglesias)
- http://www.sciencedirect.com/science/article/pii/S0022201112002388
- primarily looked for symbionts and pathological conditions
- http://www.sciencedirect.com/science/article/pii/S0022201112002388
Next Wednesday, 8;30AM, will be learning how to use C. Friedman microscope to measure different tissue sizes- looking for signs of inflammation
Still should get a better idea of what histology looks like in response to stress, become familiar with identifying tissues and noticing differences
June 28th, July 1st and 2nd 2013
Oyster HistologyLiterature Search
Sites to go back to and find full text articles and read in more detail:
- Estuarine acidification on survival and growth of Sydney rock oysters (Saccostrea glomerata)
- Immune response and mechanical stress susceptibility in Crassostrea virginica
- hemocytes and inflammation
- UW doesn't have full text access...
- http://link.springer.com/article/10.1007/s00360-011-0605-z#page-1
- Environmental prognostics: An integrated model supporting lysosomal stress responses as predictive biomarkers of animal health status
- http://www.sciencedirect.com/science/article/pii/S0141113605000681
- full: http://www.sciencedirect.com.offcampus.lib.washington.edu/science/article/pii/S0141113605000681
- lysosome accumulation (metals and chemicals)
- Survival, Blood osmolality, and gill morphology of juvenile yellow perch, rock bass, black crappie, and largemouth bass exposed to acidified soft water
- http://www.tandfonline.com/doi/abs/10.1577/1548-8659(1989)118%3C0386%3ASBOAGM%3E2.3.CO%3B2#.UdJiI_kp-So
- full: http://www.tandfonline.com.offcampus.lib.washington.edu/doi/pdf/10.1577/1548-8659%281989%29118%3C0386%3ASBOAGM%3E2.3.CO%3B2
- hyperplasia, hypertrophy, interstitial vacuolization, edema, necrosis, chloride cell cluster,
- Changes in gill histology of fathead minnows and yellow perch transferred to soft water or acidified soft water with particular reference to chloride cells (Leino, McCormick, Jenson)
- Histology of developing digestive system and effect of food deprivation in larval green sturgeon (Acipenser medirostris) (E. Gisbert et al)
June 27th 2013
Oyster Histology3:20-5:45 PM
I continued to catalog the tissue types contained in the rest of the slides today; a grand total of all 24 slides.
I'm not sure if the whole table will fit, it's pretty wide.
| H2.1 |
H2.10 |
H2.13 |
H2.14 |
H2.15 |
H2.16 |
H2.19 |
H2.21 |
H2.3 |
H2.6 |
H2.74 |
H2.8 |
H2.80 |
H2.82 |
H2.84 |
H2.85 |
H2.86 |
H2.87 |
H2.88 |
H2.9 |
H2.90 |
H2.93 |
H2.95 |
H2.96 |
|
| Gill |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
| Intestine |
x |
x |
x |
x |
x |
x |
x |
x |
? |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
|||
| Digestive Gland |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
|||||||
| Mantle |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
| Adductor |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
|
| Other |
Heart? |
premature sperm? |
see pic 32 |
heart? |
see pics 43&44 |
heart? Kidney? |
see pic 68… |
kidney or post esophagus? |
spermatids and oocytes?? (pic 77) |
see pics 83-84 |
||||||||||||||
| post-esophagus? |
||||||||||||||||||||||||
| Picture # |
27(10x), 90(4x), 91(10x) |
25(4x), 92(4x), |
28(4x), 93(10x) |
29(4x), 30(10x) |
31(4x), 94(10x), 95(40x), |
32(4x),96(10x), 33(4x) |
34(4x), 35(10x), 97(4x) |
36(4x), 37(4x), |
38(4x), 39(10x), 40(4x) |
41(4x), 42(4x), 43(10x) |
44(10x), 45(40x), 46(4x), 47(40x) |
48(4x) |
49(4x) |
50(4x), 51(4x) |
52(4x), 53(10x), 54(4x), 55(10x), 56(40x) |
56(10x), 57(10x) |
58(4x), 59(10x) |
60(4X), 61(10X), 62(4X), 63(10X), 64(40X) |
65(10x), 66(40x), 67(10x), |
68(4x), |
69(4x), 70(10x), 71(40x), 72(10x), |
73(4x),74(10x), 75(40x), 76(10x), 77(40x), 78(40x), 79(10x), 80(10x), 81(40x) |
82(4x), 83(10x) 84(40x) |
85(4x), 86(4x), 87(4x), 88(10x), 89(4x) |
| Notes: |
pic 27 might be mantle but looks like dense muscle, adductor muscle in visceral mass sample |
pic of what might be heart |
intestine appears torn/sliced, adductor dispersed/broken, 93 is closer of 28 |
really small sample, diffuse |
large slice in adductor muscle, intestine torn/sliced |
majority of tissue is digestive glad, potential heart tissue is on adductor sample |
generally difffuse, intestine torn/sliced, pic 36 is on adductor sample |
appears to be mostly mantle, can't tell if there is intestine or not, much of it is diffuse |
small blue dots in clusters throughout mantle |
diffuse adductor |
lots of gill and digestive gland |
pics 54-56 correlate to the same tissue/structure |
see pic for intestine? |
diffuse mantle tissue, possibly reporoductive follicles throughout |
diffuse adductor |
not much intestine, what's there is pretty torn |
spread out sample, lots of tearing |
79 is a zoomed out shot of 78. There seems to be a lot more hemocytes (or at least the small purple dots) throughout the mantle than in other slides |
possibly a good representative slide of wanted cross section |
much of intestine sliced/torn, lots of digestive gland; pic 89 is adductor w/other tissue? |
I also added the objective used for early slide photos and took a few more representative pictures. As I got to the later slides I was a little more comfortable with noticing other tissues and thinking of what they could look like, causing me to go back through slides from yesterday and there were a few similar structures. The slides seem pretty inconsistent when it comes to what kind of tissue is actually there; some of the cross sections are from different areas of each oyster and some are pretty torn/spread out.
To Do:
- Compile data in way that groups those with like tissues (AKA figure out Excel...) so we know what slides can actually be compared
- Ask Professor Roberts about access to the "eagle" data drop and the open notebook section of the wikispace
- Put the Excel file and pictures on eagle drop box (iPhone photos #25-97)
- Literature search
- Bivalve histology indicators of stress, OA, metaplasia, tissue types effected/ inflammation,
- Example histology pics of kidney, heart, and reproductive organs to see if that's what's on some of these slides
Planning on coming back into the lab in the morning, though may end up doing a literature search from home instead depending on car troubles I'm anticipating in the morning.
June 26th 2013
Oyster Histology10:30-11:10 AM; 2:15- 5:15 PM
Today was mostly fiddling with set-ups, figuring out the Google calendar for the lab, setting up the wikispace to correlate with the Roberts Lab page and beginning analyzing histology slides. I set up the iPhone we are using for a camera, which took more time than it should have but ended successfully! *Remember to turn on the camera first then mount the holster to the scope, directly mounting the lense to the iPhone ends up cutting off much of the field of view. I then organized Emma's slides in numerical order and set up an Excel file to include the tissue types I was looking for: gill, intestine, digestive glad, mantle, and adductor muscle, as well as space for other tissues, additional notes, and a corresponding picture number. About halfway through I started adding what objective the pictures correlated to and I probably should have done that from the start. This may mean going back through earlier pics and assigning objectives to have an accurate scale.
Things to think about:
If there is "mystery tissue", how likely is that to be kidney? Example histology pics indicate close proximity-- take a closer look at what that looks like
Reproductive structures-- maybe double check for oocytes and spermatids
Heart tissue may be present
Some tissue torn or sliced-- not sure how much of that is from the live oyster and how much was transferring into slides
The 17 slides I have so far:
| H2.1 |
H2.10 |
H2.13 |
H2.14 |
H2.15 |
H2.16 |
H2.19 |
H2.21 |
H2.3 |
H2.6 |
H2.74 |
H2.8 |
H2.80 |
H2.82 |
H2.84 |
H2.85 |
H2.86 |
|
| Gill |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
| Intestine |
x |
x |
x |
x |
x |
x |
x |
x |
? |
x |
x |
x |
x |
x |
x |
||
| Digestive Gland |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
||||||
| Mantle |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
| Adductor |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
x |
|
| Other |
Heart? |
premature sperm? |
see pic 32 |
heart? |
see pics 43&44 |
||||||||||||
| Picture # |
27 |
25 |
28 |
29(4x), 30(10x) |
31 |
32, 33 |
34, 35 |
36, 37, |
38(4x), 39(10x), 40(4x) |
41(4x), 42(4x), 43(10x) |
44(10x), 45(40x), 46(4x), 47(40x) |
48(4x) |
49(4x) |
50(4x), 51(4x) |
52(4x), 53(10x), 54(4x), 55(10x), 56(40x) |
56(10x), 57(10x) |
58(4x), 59(10x) |
| Notes: |
pic might be mantle, but looks like dense muscle |
pic of what might be heart |
intestine appears torn/sliced, adductor dispersed/broken |
really small sample, diffuse |
large slice in adductor muscle, intestine torn/sliced |
majority of tissue is digestive glad |
generally difffuse, intestine torn/sliced |
appears to be mostly mantle, can't tell if there is intestine or not, much of it is diffuse |
small blue dots in clusters throughout mantle |
diffuse adductor |
lots of gill and digestive gland |
pics 54-56 correlate to the same tissue/structure |
see pic for intestine? |
June 25th 2013
Oyster Histology9:00-10:25 AM
This was my first day working in the Roberts lab with Emma. I became more oriented with the lab and Emma lead me through some example histology pictures. We then looked at about 5 slides of hers and Mac's to give me a better idea of what kinds of tissues I was actually looking at. I will be primarily cataloging what types of tissues are in Emma's slides as they seem to be inconsistent in what tissues were included across the samples. The main tissues I am checking for are intestine, digestive glad, mantle, and gill-- the cross section of the oyster extending dorso-ventrally.
Other things to remember seeing: light blue dots (larger than hemocytes) might be mucous cells, spermatids can be dark and tightly clustered, oocytes may look similar to digestive tissues but without "star" shape, open digestive tubules may indicate metaplasia, signs of stress could be flooding of hemocytes